For neuroblastoma, the overall 5-year survival rate remains <40% (Eposito et al, 2017). In another report, a moderate increase in the survival rates (52% for 1975-77 to to 74% for 1999-2005) was documented—primarily attributed to the increased cure rate for the benign type (Maris, 2010). Nearly half of all diagnosed are classified as ‘high risk’ group for recurrence. Depending on the type of risk group to which a neuroblastoma patient is assigned, different treatment may follow. For the ‘low risk’ group with localized tumor, surgery is recommended, whereas for the ‘high risk’ group involving metastasis to bone (or bone marrow), treatments may include surgery, dose-intensive chemotherapy with hematopoietic stem cell transplantation, radiotherapy and immunotherapy. As such, the ability to diagnose accurately is critical as it directly impacts risk stratification as well as therapy.
In trying to classify patients into different risk groups (low, high, intermediate, ultra-high), multiple prognostic factors are considered, which include the age of patient, clinical stage, tumor differentiation, and tumor histology. To help identify the stage, ‘Omics’ data (i.e. proteomics, genomics, metabolomics) are increasingly being utilized. The ability to sequence the entire genome has provided insight on additional layer of information, ex. gene copy number variation, amplification, deletion, genetic variant (ex. single nucleotide polymorphism). Through high-throughput Omics analysis, other relevant information such as mRNA/protein expression level, post-translational modification and metabolites can be acquired.
Neuroblastoma is thought to be a disease of developing tissues that arise from the precursor cells (Hoehner et al, 1996). Genetic analysis has shown that hereditary neuroblastoma, which exhibits autosomal dominant mode of inheritance, is associated with the activating mutation in ALK (ana-plastic lymphoma kinase) oncogene (Mossé et al., 2008) and inactivation of homeobox gene PHOX2B (Trochet et al., 2004). Genome wide association study (GWAS) has implicated several other genes (ex. FLJ22536, BARD1). In ~20% of the cases, N-myc gene is amplified, which is associated with advanced stage neuroblastoma (Brodeur et al., 1984; Lee et al., 1984). N-myc belongs to a family of human proto-oncogenes related to v-myc—i.e. c-myc, N-myc, l-myc. V-myc is an oncogene encoded by avian myelocytomatosis virus (MC29), which causes neoplasm in chickens (myelocytomas, tumors of kidney and liver). In patients with Burkitt lymphoma, a chromosomal translocation places c-myc gene under the control of immunoglobulin promoter, resulting in elevated expression.
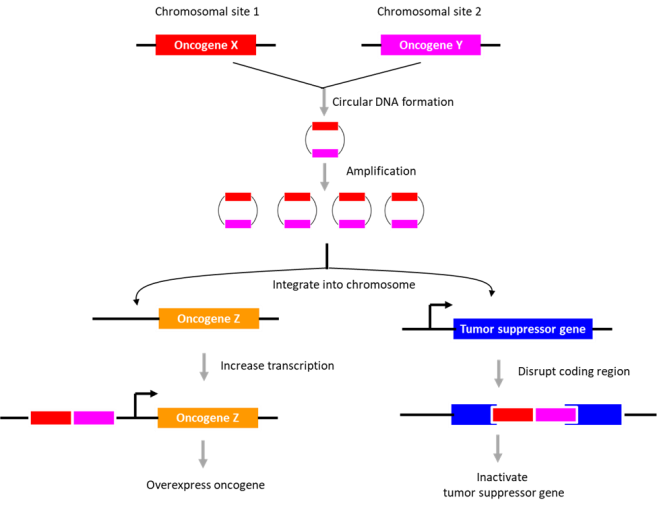
In human tissues, three different types of circular DNAs have been identified, i.e. ring chromosome, large extrachromosomal circular DNA (ecDNA), and small extrachromosomal circular DNA (eccDNA). The presence of circular
In neuroblastoma, no ring chromosomes were found although ecDNA (mean size 680 kbp; ~0.8 copies per tumor) and eccDNA (mean size 2.4 kbp; ~5600 copies per tumor) were detected. Whereas ecDNA may contain the entire genes, eccDNAs generally contains partial genes, with DNA circularization occurring in both coding and noncoding regions. Intriguingly, the chromosomal region encompassing N-myc gene was highly circularized; other genes circularized include proto-oncogene JUN or MDM2 and transcription factor SOX11 or TAL2.
The potential role of DNA circularization in tumorigenesis was investigated. Examination of circle junction sequence revealed microhomologies (minimally 5 bp), indicating that the DNA circles may have arisen during ‘microhomology-mediated DNA repair’. The data suggested that DNA circularization may be necessary but not sufficient for genomic amplification. Also, its role in gene overexpression appeared unlikely. As an alternative, its role in genome remodeling was examined. The extrachromosomal circular DNAs were found to be chimeras consisting of genomic sequences derived from distinct chromosomes (~2.2 chimeric segments for eccDNA; ~4.8 chimeric segments for ecDNA). Further, they described an eccDNA (containing a region in chromosome 2 encompassing the N-myc gene), which is partly integrated into chromosome 13, disrupting DLCK1 gene. Thus, the authors suggest that the extrachromosomal circular DNAs may integrate into various sites in the genome to disrupt tumor suppressor gene function or enhance proto-oncogene expression. The presence of “circle-derived rearrangements” correlated with poor survivability, indicative of its potential use as a diagnostic marker for neuroblastoma.
https://www.biosyn.com/oligonucleotide-modification-services.aspx
References
Brodeur GM, Seeger RC, Schwab M, Varmus HE, Bishop JM. Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. (1984) Science. 224:1121–4. PMID: 6719137 DOI: 10.1126/science.6719137
Esposito MR, Aveic S, Seydel A, Tonini GP. Neuroblastoma treatment in the post-genomic era. (2017). J Biomed Sci. 24:14. PMID: 28178969 doi: 10.1186/s12929-017-0319-y
Hoehner JC, Gestblom C, Hedborg F, Sandstedt B, Olsen L, Pahlman S. A developmental model of neuroblastoma: differentiating stroma-poor tumors’ progress along an extra-adrenal chromaffin lineage. (1996) Lab Invest. 75:659–75. PMID: 8941212
Koche RP, Rodriguez-Fos E, Helmsauer K, Burkert M, MacArthur IC, Maag J, et al. Extrachromosomal circular DNA drives oncogenic genome remodeling in neuroblastoma. (2019) Nat Genet. PMID: 31844324 DOI: 10.1038/s41588-019-0547-z
Lee WH, Murphree AL, Benedict WF. Expression and amplification of the N-myc gene in primary retinoblastoma. (1984) Nature 309:458-60. PMID: 6728001 DOI: 10.1038/309458a0
Maris JM. Recent advances in neuroblastoma. (2010) N Engl J Med. 362:2202-11. PMID: 20558371 doi: 10.1056/NEJMra0804577
Mossé YP, Laudenslager M, Longo L, et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. (2008) 455:930–5. PMID: 18724359 PMCID: PMC2672043 DOI: 10.1038/nature07261
Trochet D, Bourdeaut F, Janoueix-Lerosey I, et al. Germ-line mutations of the paired-like homeobox 2B (PHOX2B) gene in neuroblastoma. (2004) Am J Hum Genet. 74:761–4. PMID: 15024693 PMCID: PMC1181953 DOI: 10.1086/383253