The surface spike (S) protein is essential for coronavirus binding and entry into host cells. Molecular biology studies indicate that the heptad repeats 1 and 2 (HR1 and HR2) in the S protein induce the fusion of the viral membrane with the host's cell membrane. Therefore, inhibiting SARS-CoV-2 membrane fusion will potentially allow the prevention of COVID-19 infections or even treat it.
What do antiviral peptides do?
Antiviral blocking peptides targeting the viral fusion core can inhibit viral membrane fusion, thereby inhibiting the virus's entry into the host cell.
Presently we do not understand how SARS-CoV-2 enters a host cell in detail. However, models for the fusion core formation are available. The S protein is part of the viral capsid and contains two subunits S1 and S2. The S1 subunit contains the receptor-binding domain (RBD). Subunit S2 mediates the fusion between the virus and the host cell and cell entry. When the SARS-CoV-2 virus particle encounters the host cell, the RBD as part of S1 binds to the host's angiotensin-converting enzyme 2 (ACE2) receptor protein. Furin, an enzyme present in human cells, is thought to cleave the S protein into S1 and S2 subunits. This cleavage event exposes the fusion peptide (FP) of S2 and inserts it into the target cell membrane. The assembly of three HR1 and three HR2 domains form the fusion core by pulling on the cell membrane allowing fusion with the host cell membrane allowing entry of the virus into the cell.
An antiviral peptide designed to bind more tightly to the HR1 domain prevents the formation of the fusion core, and the virus cannot enter the host cell. Hence infection of the host cell is prevented.
In the spring of 2020, Xia et al. reported the X-ray crystal structure of a six-helical bundle (6-HB) core of the HR1 and HR2 domains present in the S2 subunit of the SARS-CoV-2 spike protein. Several mutated amino acid residues present in the SARS-CoV-2 HR1 sequence appear to allow enhanced interactions with the HR2 domain. A SARS-CoV-2 spike (S) protein-mediated cell–cell fusion assay revealed that SARS-CoV-2 has a superior plasma membrane fusion capacity compared to that of SARS-CoV. Xia et al. designed a series of lipopeptides derived from EK1. The study identified EK1C4 as the best inhibitor in preventing S protein-mediated membrane fusion and pseudovirus infection. EK1C4 also inhibited membrane fusion and infection of other human coronavirus and pseudoviruses. EK1C4 inhibited SARS-CoV, MERS-CoV, SARSr-CoVs, five live human coronaviruses, and SARS-CoV-2.
Intranasal application of EK1C4 before or after challenge with HCoV-OC43 protected mice from infection. These findings suggest that EK1C4 potentially prevents infections by the currently circulating SARS-CoV-2 and other emerging SARS coronaviruses. It is also possible to use EK1C4 for the treatment of COVID-19.
A second study performed by Ling et al. using in-silico analysis predicted the HR1 and HR2 regions in the S protein via sequence alignments. A computational model allowed modeling the binding energies of HR1 and HR2 of the fusion core. The model guided the design of antiviral peptides.
The study identified a homologous sequence region in the heptad repeats (HR) region of SARS-CoV-2 and SARS-CoV. Molecular dynamics-based modeling allowed the design of an antiviral peptide that potentially prevents virus membrane fusion with the host cell membrane. The study reported calculated binding energies for HR1 and HR2 in the SARS-CoV-2 spike protein as well.
Table 1: Antiviral Peptides
Peptide | Sequence | Reference |
|
|
|
EK1 | SLDQINVTFLDLEYEMKKLEEAIKKLEESYIDLKEL | Xia et al. |
EK1C4 | SLDQINVTFLDLEYEMKKLEEAIKKLEESYIDLKELGSGSG-PEG4-cholesterol | Xia et al. |
Scramble | LKVLLYEEFKLLESLIMEILEYQKDSDIKENAEDTK | Xia et al. |
|
|
|
Linker Peptide | SGGRGG | Xia et al. |
|
|
|
HR1 | NQKLIANQFNSAIGKIQDSLSSTASALGKLQDVVNQNAQALNTLVKQ | Ling et al. |
HR2 | GINASVVNIQKEIDRLNEVAKNLNESLIDL | Ling et al. |
Linker Peptide | LVPRGSGGSGGSGGLEVLFQGP | Ling et al. |
Fusion Peptide | HR1-Linker-HR2 | Ling et al. |
|
|
|
HR2 antiviral | DISGINASVVNIQKEIDRLNEVAKNLNESLIDLQEL | Ling et al. |
Other types of antiviral peptides inhibit the binding of the SARS coronavirus spike RBD to the cellular receptor, ACE2. Often these peptides are also called "Coronavirus Inhibitory Peptides."
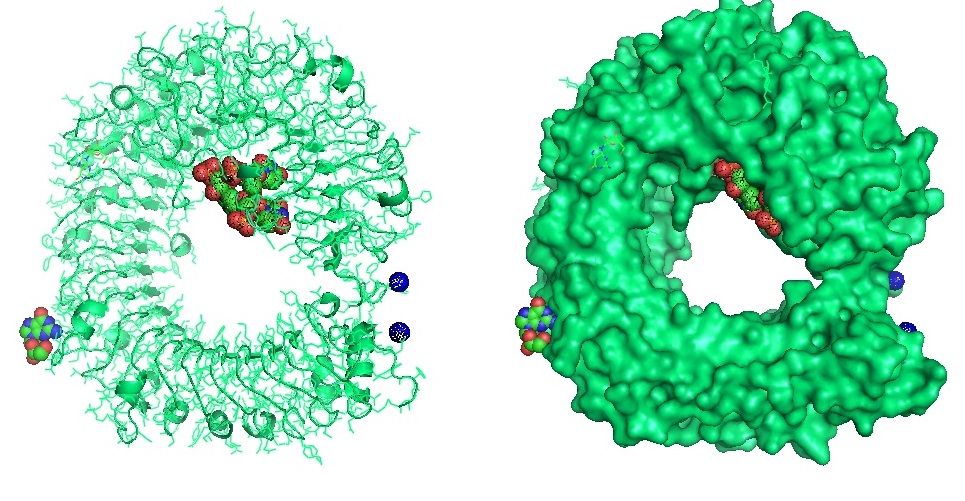
Interaction of EK1 with HCoV.
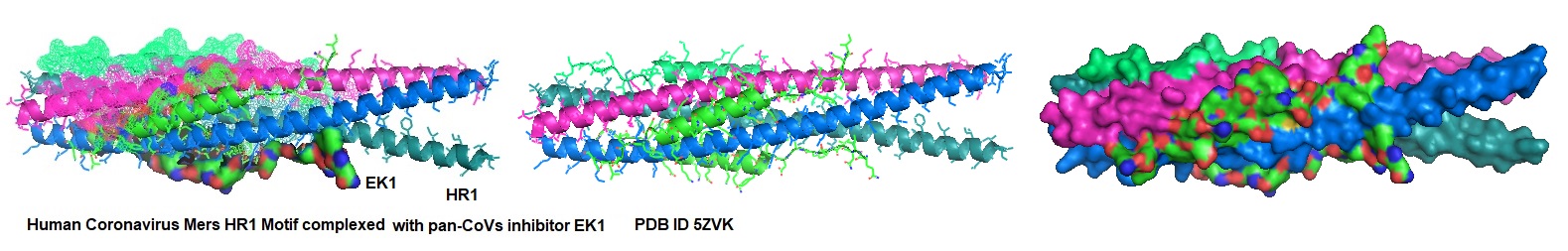
Figure 1: Interaction of EK1 with HCOV HR1. Xia et al. observed that peptide OC43-HR2P, derived from the HR2 domain of HCoV-OC43, exhibited broad fusion inhibitory activity against multiple human coronaviruses (HCoVs). The optimized peptide EK1 showed substantially improved pan-CoV fusion inhibitory activity and pharmaceutical properties. The crystal structures indicated that EK1 could form a stable six-helix bundle structure with short α-HCoV and long β-HCoV HR1s. This finding supported the role of the HR1 region as a viable pan-CoV target site further. The structural models illustrate that EK1 snugly fits into the hydrophobic grooves formed between two adjacent HR1 helices of the 3HR1 core from MERS-CoV, SARS-CoV, and 229E. The EK1 peptide is shown as a green ribbon.
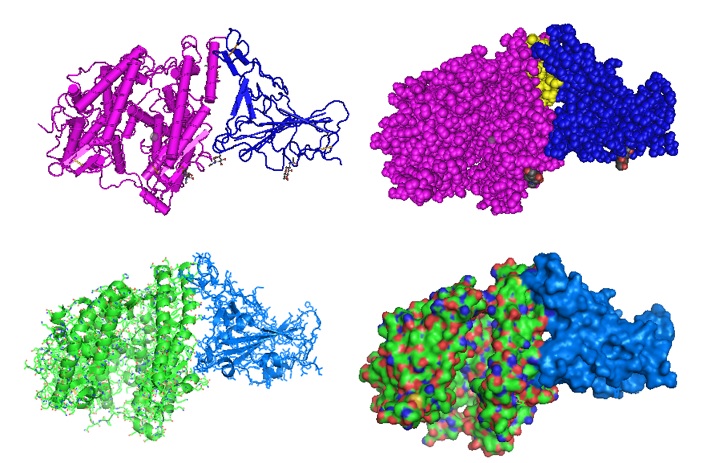
Interaction of HR peptides illustrating the post fusion core.

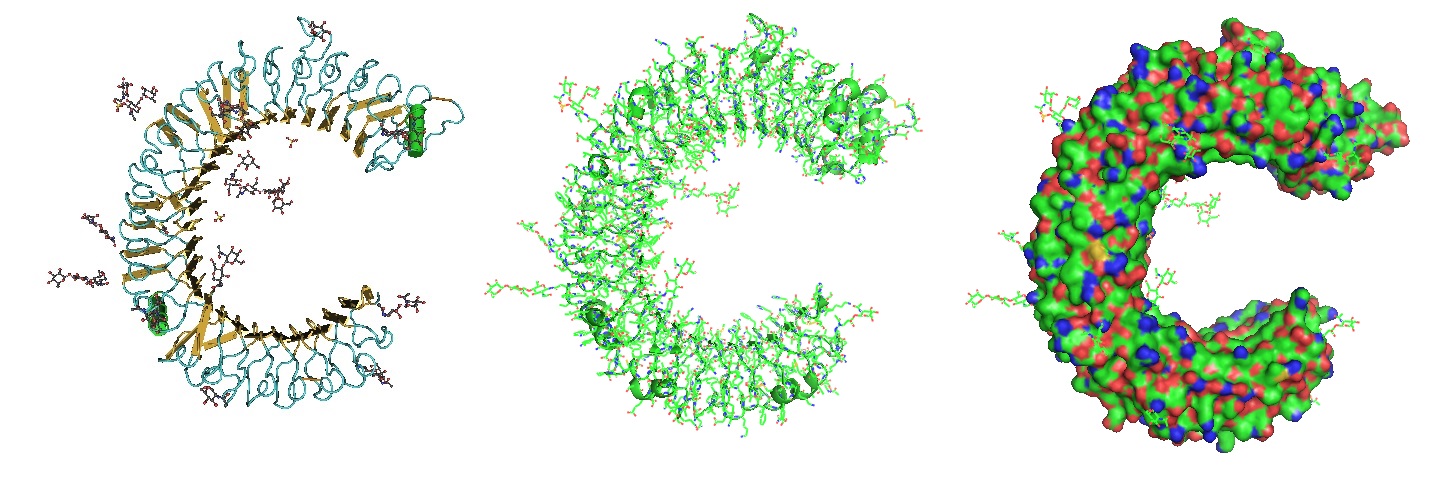
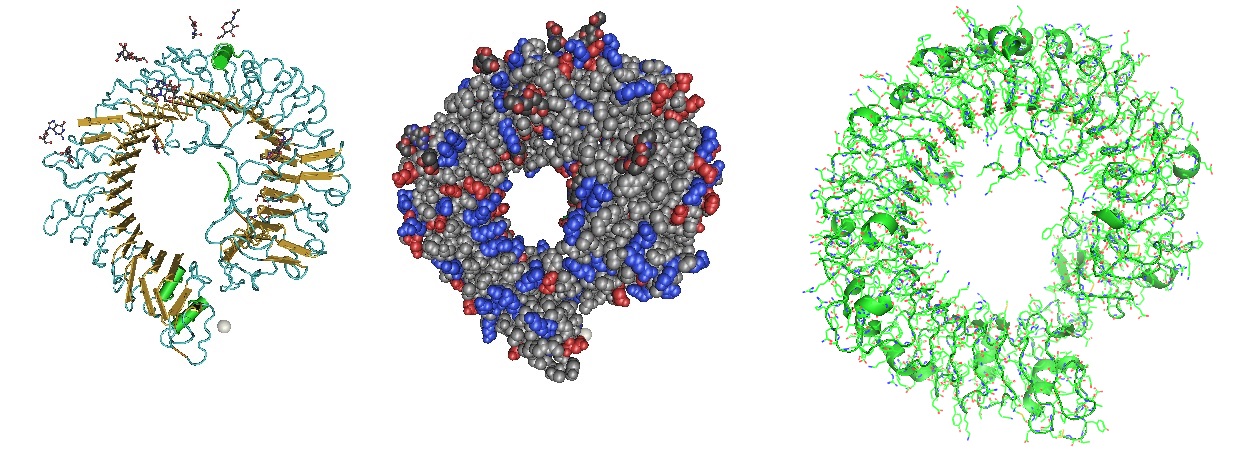
Figure 2: Model of the HR1/HR2 complex. The favored model of virus entry into the host cell assumes that furin cleaves the S protein into the S1 subunit and the S2 subunit. The cleavage exposes the fusion peptide (FP) of S2 and inserts it into the target cell membrane. Three HR1s and three HR2s combine to form the fusion core, pulling the viral membrane to fuse with the host cell membrane. Ling et al. suggested that the designed and computational-optimized anti-virus peptide bind to HR1 more tightly, thereby preventing the HR1s and HR2s from forming the fusion core. Hence, the fusion peptide is a candidate for an antiviral peptide.
Reference
Coronavirus Inhibitory Peptides
Rongsong Ling, Yarong DaI, Boxuan Huang, Wenjie Huang, Jianfeng Yu, Xifeng Lu, Yizhou Jiang; In silico design of antiviral peptides targeting the spike protein of SARS-CoV-2. PeptidesVolume 130, August 2020, 170328. https://www.sciencedirect.com/science/article/pii/S0196978120300772 . Published in August 2020.
.png)

.png)

.jpg)










.jpg)





.jpg)



.png)
.png)


.jpg)