Peptide-based therapies offer a cost-effective and rapidly developed therapeutic response to pandemic outbreaks such as COVID-19. However, one drawback is their proteolytic instability. Peptidomimetics, the art of designing molecules that mimic natural peptides, promises to overcome these hurdles.
For the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2; COVID-19), therapeutic peptides and peptide-based therapeutics may target main viral entry pathways into human cells. One involves the interaction between the host angiotensin-converting enzyme-2 (ACE2) receptor protein and the viral spike or surface (S) protein.
The S protein allows the coronavirus to bind to the host cell receptor ACE2. Peptides designed to interfere with the binding event can inhibit viral entry, thereby preventing infection. These type of peptides are also known as antiviral-peptides. However, to stabilize functional natural peptides peptide mimetics will need to be designed.

Novel Coronavirus SARS-CoV-2: This image shows SARS-CoV-2 (round blue objects) emerging from the surface of cells cultured in the lab. A scanning electron microscope was used for its generation. The corona virus SARS-CoV-2, or 2019-nCoV, is the cause of COVID-19. The virus shown was isolated from a patient in the U.S. The image was captured and colorized at NIAID's Rocky Mountain Laboratories (RML) in Hamilton, Montana.
{ Credit: NIAID-RML) TEMI Ref: Prasad S, Potdar V, Cherian S, Abraham P, Basu A; ICMR-NIV NIC Team. Transmission electron microscopy imaging of SARS-CoV-2. Indian J Med Res. 2020 Feb & Mar;151(2 & 3):241-243. doi: 10.4103/ijmr.IJMR_577_20 [PMC]. }
Coronavirus SARS-CoV-2 viroid and genome

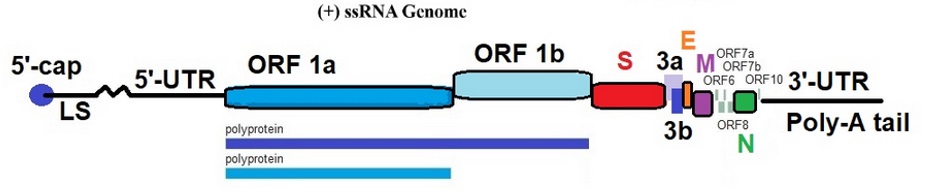
Virion: Enveloped, spherical, 60 to 140 nm in diameter with 9 to 12 nm spikes
Genome: ~ 30 kb positive-sense, ssRNA
RNA Transcript: 5'-cap, 3'-poly-A tail
Proteome: ~ 10 proteins
Transmission: Links to seafood and animal market cases suggest animal-to-human transmission.
Sustained human-to-human transmission observed in later cases.
Phylogeny: Closely related to bat-SL-CoVZC45 and bat-SL-CoVZX21.
The typical organization of the viral genome is
5’-leader-UTR-replicase-S(Spike)-E(Envelope)-M(Membrane)-N(Nucleocapsid)-3’-UTR-poly(A) tail.
Accessory genes are interspersed within the structural genes at the 3’-end of the genome.
Structural models of SARS-CoV-2 S protein

The angiotensin-converting enzyme 2 (ACE2) is the cellular receptor for the SARS coronavirus (SARS-Cov or SARS-CoV-1) and SARS-CoV-2 S protein. The following peptide is at the interphase between the ACE2 receptor protein and the receptor binding domain (RBD) of the SARS-CoV-2 S protein.
hACE2 n-terminal peptide AA30 to 53:IEEQAKTFLDKFNHEAEDLFYQSS
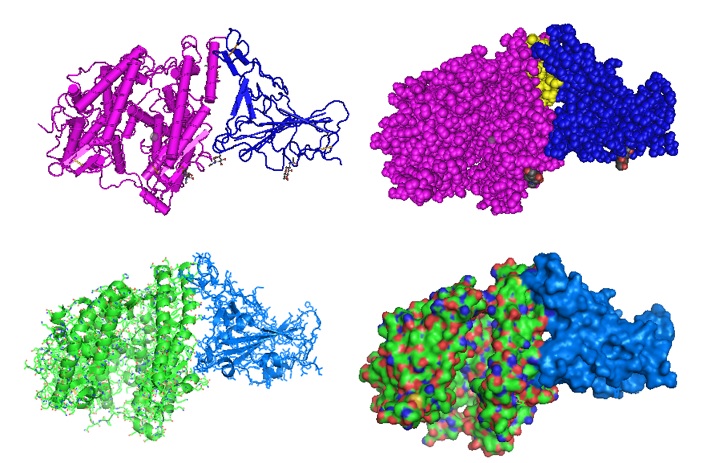
Figure 1: Structural model of the SARS-Cov-2 RBD-ACE2 complex [PID 6LZG]. The location of the inter-phasing peptide is indicated in yellow. The crystal structure of the C-terminal domain (RBD) of SARS-CoV-2 (SARS-CoV-2-CTD or SARS-Cov-2 RBD) as part of the spike (S) protein in complex with human ACE2 (hACE2) was solved by Wang et al. in 2020. The structural model reveals how human angiotensin hACE2 binds and interacts with the S protein in a mode similar to that observed for SARS-CoV.
{ Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, Lu G, Qiao C, Hu Y, Yuen KY, Wang Q, Zhou H, Yan J, Qi J. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell. 2020 May 14;181(4):894-904.e9. [PMC] }
SARS-CoV-2 S protein RBD interphasing peptide
This S protein RBD interphase peptide is in contact with the ACE2 receptor protein.
SARS-CoV-2 peptide AA483 to 506: VEGFNCYFPLQSYGFQPTNGVGYQ
.jpg)
Figure 2: CryoEM structure of the SARS-CoV-2 Spike protein in complex with ACE2 (PID 7A96): (Left image) The location of the ACE2 peptide IEEQAKTFLDKFNHEAEDLFYQSS on the interphase with the S protein in the complex is shown in yellow. (Right image) The S protein RBD peptide VEGFNCYFPLQSYGFQPTNGVGYQ in contact with ACE2 in the complex is shown in yellow. The structure as solved by Benton et al. revealed a refolding event of the S1 subdomain after binding to ACE2. The refolding disrupts interactions with the S2 subdomain which destabilized the structure of S2 proximal to the secondary (S2') cleavage site.
{ Benton DJ, Wrobel AG, Xu P, Roustan C, Martin SR, Rosenthal PB, Skehel JJ, Gamblin SJ. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature. 2020 Sep 17. doi: 10.1038/s41586-020-2772-0. Epub ahead of print. PMID: 32942285. [PMC]. }
---...---