The recent COVID-19 pandemic has rapidly expanded to infect more than 14 million people by July 21, 2020, and has had a significant impact on our lives. The innate immune system is one of the first lines of defense against pathogens, including viruses. The new coronavirus called severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the cause of COVID-19 infections. SARS-CoV-2 emerged in China in late 2019 in the Wuhan region. COVID-19 has been declared a pandemic by the World Health Organization in March 2020.
Recently Qiu et al. developed a biosensor for the selective detections of SARS-CoV-2 nucleic acids. The research group designed a dual-functional localized surface plasmon resonance (LSPR) based biosensor by combining the photothermal effect with plasmonic sensing transduction for SARS-CoV-2 viral nucleic acid detection. The new plasmonic chip contains a two-dimensional distribution of nanoabsorbers (AuNIs). The chip generates a local plasmonic photothermal heat (PPT) transduced in situ hybridization, allowing a highly sensitive and accurate SARS-CoV-2 detection.
Cells, both prokaryotic and eukaryotic, also have mechanisms for sensing RNA, for example the entry of foreign RNA into cells. Human epithelial cells contain RNA sensing molecules outside and inside the cell body. Epithelial cells are at the interface where self and non-self meets. In humans, in the respiratory tract, these cells are the front-line physical barrier between the environment and the body. The respiratory system is integral to innate and adaptive immunity. The epithelium plays an important role in host defense. Epithelium cells secrete a wide spectrum of protective, antimicrobial, homeostatic, and immune regulatory factors. Secreted molecules include antioxidants, defensins, collectins, complement components, lactoferrin, lysozyme, and protease inhibitors. In humans, oral epithelial cells line the oral cavity and are an effective and dynamic barrier to invading pathogens. Epithelial cells contain an extensive repertoire of innate defenses.
In 2012, Ghosh et al. studied the proteome of healthy human oral epithelial cells. The use of two-dimensional electrophoresis followed by liquid chromatography on-line with tandem mass spectrometry (2DE-LC-MS/MS) and database searches allowed the identification of 237 most abundant proteins present in oral epithelial cells. Nucleic acid-binding proteins (20.4%), transferases (11%), cytoskeletal proteins (7.6%), and enzyme modulators (7.2%) are the three most abundant protein classes identified. Also, the study identified hydrolases, isomerases, phosphatases, proteases, transcription factors, and chaperones.
Querying the innate database (innateDB), the research group identified 64 proteins associated with innate immunity. The 64 observed proteins associated with innate immunity confirmed that oral tissue acts as a primary active barrier to infection.
RNA sensors in the cytosol contain three members of the RIG-I-like receptors (RLRs). These are the retinoic acid-inducible gene I (RIG-I), the melanoma differentiation factor 5 (MDA5), and the laboratory of genetics and physiology 2 (LGP2). The three proteins share a similar organization. These proteins contain three distinct domains:
(i) a C-terminal repressor domain (RD) embedded within the C-terminal domain (CTD);
(ii) a central ATPase containing DExD/H-box helicase domain that can bind RNA; and
(iii) a N-terminal tandem CARD domain that mediates downstream signaling.
Each retinoic acid-inducible gene I like receptor senses different viral infections. Different replicating viral RNA are recognized by the receptors in the cytoplasm. The Retinoic Acid Inducible Gene-I (RIG-I) detects pathogenic RNAs carrying a 5′-triphosphate (5′ppp). RIG-I, when activated, triggers a Type I interferon (IFN) response. RIG-I does not recognize self RNAs such as mRNAs because they are capped post-transcriptionally on the 5′-end with 7-methyl guanosine (m7G) and 2′-O-methylation of 5′-end nucleotides. Viruses have evolved mechanisms to mimic these modifications for evasion of the innate immune response. Studies performed by Devakar et l. provide structural and mechanistic insights into the roles of the m7G cap and 2′-O-methylation in RIG-I evasion. They also showed that RIG-I accommodates the m7G base while maintaining 5′ppp contacts and can recognize Cap-0 RNAs but not Cap-1. Cap-0 and 5′ppp double-stranded (ds) RNAs bind to RIG-I with nearly identical Kd values and activate RIG-I’s ATPase and cellular signaling response to similar extents.
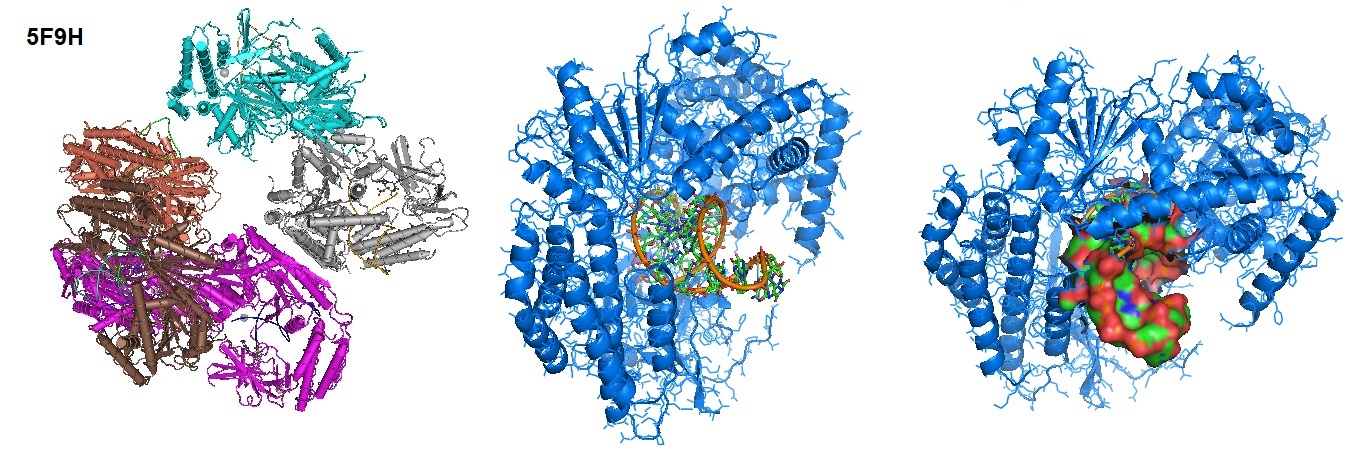
Figure 1: Structure of RIG-I Helicase-Repressor Domain in complex with 5'-ppp hairpin (HP) RNA.
The CARD domain is present in RIG-I and MDA5 but absent in LGP2. The activation by RNA ligands recruits RIG-I and MDA5 to the adaptor protein Mitochondrial Antiviral Signaling (MAVS) via a CARD–CARD, which in turn activates NF-κB and IRFs.
Toll-like receptors (TLRs) are receptors of the innate immune system that recognize conserved “pathogen-associated molecular patterns” of microbes and viruses. Activation of TLRs establishes an antiviral immune response. The antiviral immune response coordinates long-lasting adaptive immunity to control viral infection. However, after microbe-induced damage to the host tissues, “danger-associated molecular patterns” that also activate TLRs can lead to an overreacting inflammatory response, which ultimately leads to tissue damage.
The new SARS-CoV2 coronavirus is a positive-sense single-stranded RNA virus. In vitro experiments on spike proteins of the virus indicated affinity with the angiotensin-converting enzyme receptor 2 (ACE2). ACE2 appears to be the primary receptor protein allowing host cell entry of the virus. Following cell entry, the virus distributes through the circulatory stream. In some patients, viral infection triggers a systemic response leading to hyper inflammation. The hyperactivation of the immune system occurring in some infected people appears to significantly damage the lung, ultimately leading to the patients’ death. During the entry of host cells by the coronavirus SARS-CoV-2, the virus trimeric spike glycoprotein interacts with its cellular receptor angiotensin-converting enzyme 2 (ACE2). Host proteases, for example, cathepsins, furin, or members of the type II transmembrane serine proteases (TTSP) family, such as Transmembrane protease serine 2 (TMPRSS2), are thought to enable virus entry by proteolytically activating virus ligands.
The innate immune system uses Germ-line-encoded pattern recognition receptor proteins (PRRs). A variety of cells express PRRs. These cells are responsible for sensing the presence of pathogen invasions. During infection, Toll-Like Receptor (TLR) family members upregulate anti-viral and pro-inflammatory mediators. Pro-inflammatory mediators include interleukin (IL)-6, IL-8, type I and type III Interferons, and others. Activation occurs through Nuclear Factor (NF)-kB. In some cases, when the cellular entry of the virus and innate immune responses are uncontrolled, a deleterious systemic response can sometimes occur in infected patients. These molecular events can lead to a “cytokine storm.” The downregulation of dendritic cells, macrophages, and T-cell functions initiate multiple organ failure, ultimately leading to death.
According to Li et al., the highest ACE2 expression levels are found in the small intestine, testis, kidneys, heart, thyroid, and adipose tissue. Lowest levels are observed in the blood, spleen, bone marrow, brain, blood vessels, and muscle. Medium expression levels of ACE2 are found in the lungs, colon, liver, bladder, and adrenal gland.
Humans are exposed to millions of pathogens daily. The adaptive immune system helps us to avoid infections. However, adaptive immune responses are slow to develop after exposure to a new pathogen. To respond, specific clones of B and T cells have to become activated and expand. Therefore, it can take a week or longer before the responses are effective. During the first critical hours and days after we are exposed to a new pathogen, our body relies on the innate immune system to protect us from infection. The innate immune system is one of the two immune systems found in vertebrates, including in humans.
Innate immunity is a nonspecific defense mechanism reacting to an antigen in the human body within hours of its appearance. Defense mechanisms of the innate immune system include physical barriers such as skin, chemicals in the blood, and immune system cells that attack foreign cells. However, innate immune responses are not specific to pathogens in the same way that adaptive immune responses are. Vertebrates require innate immune responses for the activation of the adaptive immune responses.
The skin and epithelial surfaces and tissue cells forming the outer layer of the human body and layers of cells that line hollow organs and glands provide a physical barrier between the body and the outside world. A mucus layer covers interior surfaces. The slimy mucus primarily containing mucin and other glycoproteins help prevent pathogens from adhering to the epithelium, the tissue forming the outer layer of the body surface. Also, the mucus layer contains substances that kill pathogens or inhibit their growth, such as defensins. Defensins are small cysteine-rich cationic peptides or proteins typically 12 to 50 amino acids in length produced by the innate immune system. Defensins kill phagocytosed pathogens. However, it is still unclear how exactly they do this. The thought is that they insert themselves into the cell membrane of their victims by disrupting membrane integrity.
A cell of the innate immune system recognizes pathogen-associated molecules through germline-encoded pattern recognition receptors (PRRs) present on the cell surface or within distinct intracellular compartments. PRRs identify viral pathogens by engaging pathogen-associated molecular patterns (PAMPs). These include the Toll-like receptors (TLRs), the retinoic acid-inducible gene I-like receptors (RLRs), the nucleotide oligomerization domain-like receptors (NLRs, also called NACHT, LRR and PYD domain proteins) and cytosolic DNA sensors.
The TLR family is a crucial component of mammalian immunity and acts as an early surveillance mechanism of infections. TLRs sense infection via pattern recognition of specific molecules. TLRs are proteins involved in the development and activation of innate immunity. The toll-like receptor family of 11 transmembrane receptor proteins recognizes pathogen-associated molecular patterns (PAMPs). Different types of ligands originating from a variety of pathogens activate TLRs.
Coronaviruses appear to trigger a cytokine release in the human body. The release of primarily interleukin 6 (Il-6) and other proteins of the acute phase activates the immune response.
Often homo- or heterodimers of TLR help clear infections. Some TLR types are expressed on the cell surface. Others are present on intracellular membranes of endosomes (TLR3, 7, 8, and 9). TLRs react to viral infections by detecting foreign types of nucleic acids. In healthy immune cells and epithelial cells, TLR 1, 2, 4, 5, 6, and 10 are usually expressed on the cell surface. However, TLR 3, 7, 8, and 9 are mainly expressed on endosomes, lysosomes, and endoplasmic reticulum surfaces. Coronaviruses contain single-stranded RNA (ssRNA). TLR3 and 9 interact with double-stranded viral RNA and unmethylated CpG DNA from bacteria and viruses, respectively. TLR3 is thought to respond to infections by the West Nile virus in a rodent model. The West Nile virus is a single-stranded virus.
TLR3, -7, and -8 found in the intracellular compartments recognize nucleic acid motifs. Intercellular compartments are found in the endoplasmic reticulum (ER), endosomes, lysosomes, and endolysosomes. .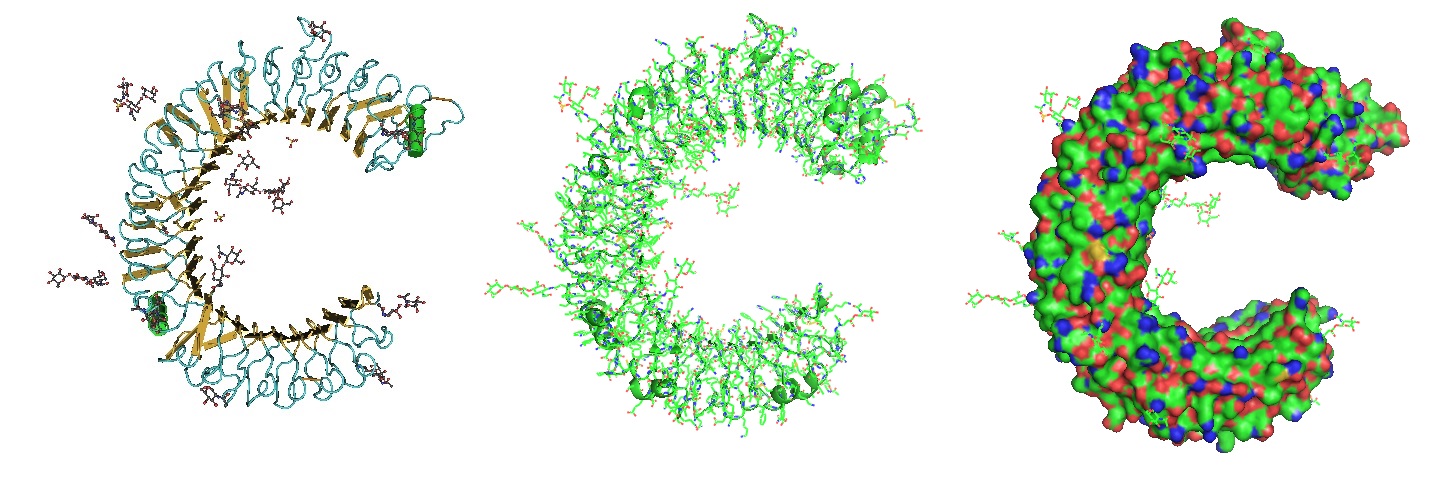
Figure 2: Structure of the Toll Like Receptor 3 Ectodomain (TLR3-ECD). Three renderings of the TlR3-ECD crystal structure at a resolution of 2.4 Å solved by Bell et al. in 2005 is shown. TLR3 senses a variety of ligands ranging from lipopolysaccharide to double stranded RNA through the ligand-binding domain composed of leucin-rich repeats. The N-linked glycans are shown as green ball-and-stick (Bell et al. 2005).
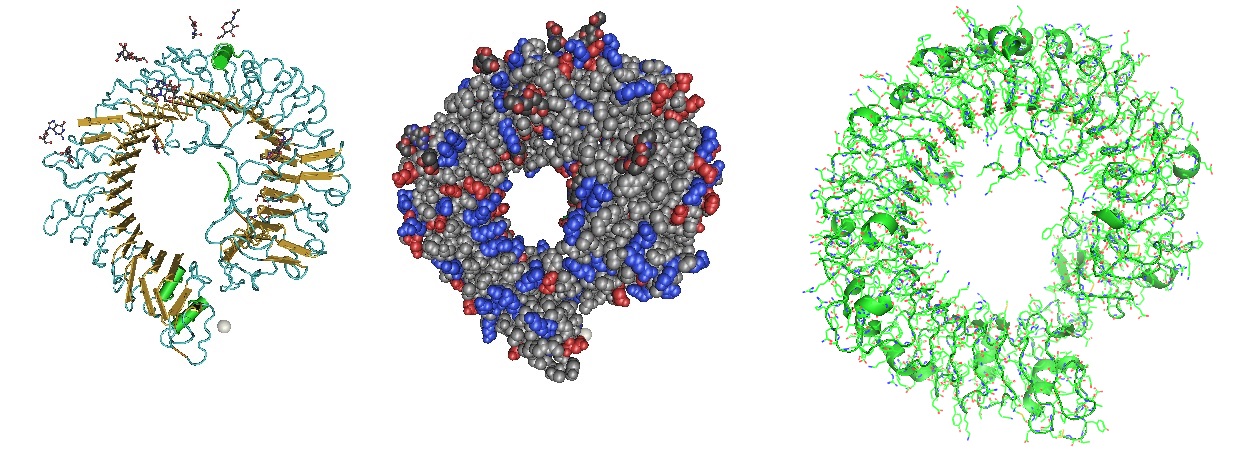
Figure 3: Structure of the Toll Like Receptor 7 (Zhang et al. 2016). Toll-like receptor 7 (TLR7) is a single-stranded RNA (ssRNA) sensor in innate immunity. TLR7 also responds to guanosine and chemical ligands, such as imidazoquinoline compounds since it contains two binding sites. One site that binds small ligands and a second site for ssRNA. TLR7 is a sensor for guanosine and uridine-containing ssRNA and is activated by both guanosine and ssRNA.
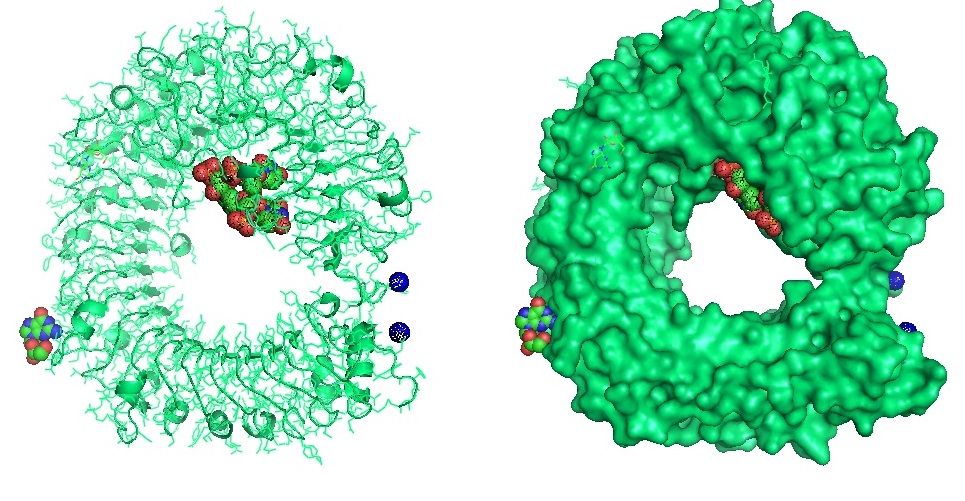
Figure 4: Structure of the Toll Like Receptor 7 with complexed with guanosine and a small uridine-containing single stranded RNA. Zhang et al. reported that the crystal structures of three TLR7 complexes form an activated m-shaped dimer with two ligand-binding sites. The first site is conserved in TLR7 and TLR8. This binding site is used for small ligand-binding and is essential for its activation. The second site differs that of TLR8 binds ssRNA. Binding to ssRNA enhances affinity of the first-site ligands. The first site preferentially recognizes guanosine and the second site specifically binds uridine moieties in ssRNA. Structural, biochemical, and mutagenesis studies revealed that TLR7 is a dual receptor for guanosine and uridine-containing ssRNA.
Table 1: Toll Like Receptors recognizing RNA
Receptor | Virus | Ligand |
TLR7 | Influenza A virus | ssRNA |
Vesicular stomatitis virus | ssRNA | |
Human immunodeficiency virus | ssRNA | |
Dengue virus | ssRNA | |
Sendai virus | ssRNA | |
Lactate dehydrogenase-elevating virus | ssRNA | |
Mouse mammary tumor virus | ssRNA | |
Murine leukemia virus | ssRNA | |
TLR8 | Human immunodeficiency virus | ssRNA |
TLR3 | Reoviridae | dsRNA |
Respiratory syncytial virus | dsRNA | |
West Nile virus | dsRNA | |
Coxsackievirus B3 | dsRNA | |
Poliovirus | dsRNA | |
Influenza A virus | dsRNA | |
Punta Toro virus | dsRNA | |
TLR2 | Measles virus | HA |
Lymphocytic choriomeningitis virus | ? | |
Hepatitis C virus | Core protein/NS3 | |
TLR4 | Respiratory syncytial virus | Fusion protein |
Coxsackievirus B4 | ? | |
Mouse mammary tumor virus | Envelope protein | |
Murine leukemia virus | Envelope protein |
Reference
Akira S, Uematsu S, Takeuchi O; 2006. Pathogen recognition and innate immunity. Cell 124:783–801.
Jessica K. Bell, Istvan Botos, Pamela R. Hall, Janine Askins, Joseph Shiloach, David M. Segal, David R. Davies; The molecular structure of the Toll-like receptor 3 ligand-binding domain. Proceedings of the National Academy of Sciences Aug 2005, 102 (31) 10976-10980; DOI: 10.1073/pnas.0505077102 . https://www.pnas.org/content/102/31/10976
Birra D, Benucci M, Landolfi L, Merchionda A, Loi G, Amato P, Licata G, Quartuccio L, Triggiani M, Moscato P. COVID 19: a clue from innate immunity. Immunol Res. 2020 Jun;68(3):161-168. doi: 10.1007/s12026-020-09137-5. PMID: 32524333; PMCID: PMC7286633. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7286633/
de Groot NG, Bontrop RE. COVID-19 pandemic: is a gender-defined dosage effect responsible for the high mortality rate among males? Immunogenetics. 2020 Jul;72(5):275-277. doi: 10.1007/s00251-020-01165-7. PMID: 32342146; PMCID: PMC7186185.
Devarkar SC, Wang C, Miller MT, et al. Structural basis for m7G recognition and 2'-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I. Proc Natl Acad Sci U S A. 2016;113(3):596-601. doi:10.1073/pnas.1515152113. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4725518/
Fitzgerald KA, Kagan JC. Toll-like receptors and the control of immunity. Cell. 2020;180(6):1044–1066. doi: 10.1016/j.cell.2020.02.041.
Ghosh SK, Yohannes E, Bebek G, Weinberg A, Jiang B, Willard B, Chance MR, Kinter MT, McCormick TS. Proteomic and bioinformatic profile of primary human oral epithelial cells. J Proteome Res. 2012 Nov 2;11(11):5492-502. doi: 10.1021/pr3007254. Epub 2012 Oct 16. PMID: 23035736; PMCID: PMC3508721. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3508721/
Guangyu Qiu, Zhibo Gai, Yile Tao, Jean Schmitt, Gerd A. Kullak-Ublick, and Jing Wang; Dual-Functional Plasmonic Photothermal Biosensors for Highly Accurate Severe Acute Respiratory Syndrome Coronavirus 2 Detection. ACS Nano 2020 14 (5), 5268-5277 . DOI: 10.1021/acsnano.0c02439. https://pubs.acs.org/doi/10.1021/acsnano.0c02439
Innate Immunity in Molecular Biology of the Cell, 4th edition. https://www.ncbi.nlm.nih.gov/books/NBK26846/
Søren Jensen, Allan Randrup Thomsen; Sensing of RNA Viruses: a Review of Innate Immune Receptors Involved in Recognizing RNA Virus Invasion. Journal of Virology Feb 2012, 86 (6) 2900-2910; DOI: 10.1128/JVI.05738-11.
Jensen S, Thomsen AR. Sensing of RNA viruses: a review of innate immune receptors involved in recognizing RNA virus invasion. J Virol. 2012 Mar;86(6):2900-10. doi: 10.1128/JVI.05738-11. Epub 2012 Jan 18. PMID: 22258243; PMCID: PMC3302314.
Jouhi L, Renkonen S, Atula T, Mäkitie A, Haglund C, Hagström J. Different Toll-Like Receptor Expression Patterns in Progression toward Cancer. Front Immunol. 2014 Dec 15;5:638. doi: 10.3389/fimmu.2014.00638. PMID: 25566251; PMCID: PMC4266018.
Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor [published online ahead of print, 2020 Mar 4]. Cell. 2020;S0092–8674(20)30229–4. doi:10.1016/j.cell.2020.02.052
Khan S, Siddique R, Shereen MA, et al. The emergence of a novel coronavirus (SARS-CoV-2), their biology and therapeutic options [published online ahead of print, 2020 Mar 11]. J Clin Microbiol. 2020;JCM.00187–20. doi:10.1128/JCM.00187-20.
Kumar H, Kawai T, Akira S. Toll-like receptors and innate immunity. Biochem Biophys Res Commun (2009) 388(4):621–5.10.1016/j.bbrc.2009.08.062 [PubMed] [CrossRef] [Google Scholar]
Li, M., Li, L., Zhang, Y. et al. Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infect Dis Poverty 9, 45 (2020). https://doi.org/10.1186/s40249-020-00662-x
Thompson MR, Kaminski JJ, Kurt-Jones EA, Fitzgerald KA. Pattern recognition receptors and the innate immune response to viral infection. Viruses. 2011 Jun;3(6):920-40. doi: 10.3390/v3060920. Epub 2011 Jun 23. PMID: 21994762; PMCID: PMC3186011.
Parker LC, Prince LR, Sabroe I. Translational mini-review series on Toll-like receptors: networks regulated by Toll-like receptors mediate innate and adaptive immunity. Clin Exp Immunol. 2007 Feb;147(2):199-207. doi: 10.1111/j.1365-2249.2006.03203.x. PMID: 17223959; PMCID: PMC1810480. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1810480/
Sallenave JM, Guillot L. Innate Immune Signaling and Proteolytic Pathways in the Resolution or Exacerbation of SARS-CoV-2 in Covid-19: Key Therapeutic Targets? Front Immunol. 2020 May 28;11:1229. doi: 10.3389/fimmu.2020.01229. PMID: 32574272; PMCID: PMC7270404.
Wang T, Town T, Alexopoulou L, Anderson JF, Fikrig E, Flavell RA. Toll-like receptor 3 mediates West Nile virus entry into the brain causing lethal encephalitis. Nat Med. 2004;10:1366–1373. doi: 10.1038/nm1140. [PubMed] [CrossRef] [Google Scholar]
Zhang Z, Ohto U, Shibata T, et al. Structural Analysis Reveals that Toll-like Receptor 7 Is a Dual Receptor for Guanosine and Single-Stranded RNA. Immunity. 2016;45(4):737-748. doi:10.1016/j.immuni.2016.09.011 . https://pubmed.ncbi.nlm.nih.gov/27742543/
---...---






![Robert Priest - People Like You and Me (2024) [Hi-Res]](http://www.dibpic.com/uploads/posts/2025-07/1751805912_lhttl01vuwv8a_600.jpg)








