The taste of sugars amino acids, peptides, nucleotides, and proteins

Taste and smell are chemical senses that give us humans and many animals information about the chemical composition that surround us and are part of our nearest environment.
These types of sensations are produced by a stimulating chemical that came in contact with and stimulated our gustatory nerve endings in tongues taste buds. Gustatory nerves are sensory nerve fibers innervating the taste buds and are associated with taste. However, how an individual perceives taste involves many factors. Scientists now know that how we perceive taste is influences by gene combinations coding for individual taste receptors that an individual possesses and the environment she or he lives or grew up in. Each person’s genetic makeup influences the configuration of taste receptors of the individual and determines how she or he detects the basic tastes. Our genes play a part in determining our taste preference and our environment can influence how we learn to experience new tastes. For example one person may like the taste of broccoli or chocolate better than another. The possibility of the presence of different genes encoding for different bitter receptors in different persons explains why some people find that broccoli tasted bitter and others do not. In addition, some tastes can be acquired. A persons experience is also important in determining food preferences. We all know that infants and young children need to learn what foods are safe to eat. Even before a child is born, information about specific flavors of the mothers’ diets can pass to the infant through amniotic fluid. Mammals have taste receptors which gives them the ability to distinguish different chemical substances by taste. This ability benefits their feeding behavior by helping the organism to select favorable and to avoid noxious or toxic substances. A sweet taste usually indicates desirable carbohydrate contents, whereas a bitter taste is associated with toxic substances such as alkaloids.
Humans have the ability to distinguish five types of taste – sweet, sour, salt, bitter and umami. The hydrogen ions in acids produce a sour taste whereas sodium ions from water soluble salts taste salty. However, bitter, sweet, and umami tastes are mediated through G protein coupled receptor signaling pathway systems.
The human olfactory system has a rich evolutionary history. Unlike other mammals humans are less dependent on chemosensory input but olfactory functions still play a critical role in health and behavior. The olfactory system is involved in diverse physiologic processes such as the detection of hazards in the environment, generating feelings of pleasure, promoting adequate nutrition, influencing sexuality, and maintenance of mood, pheromone detection, mother–infant bonding, food preferences, kin recognition and mating, in central nervous system physiology, and even longevity.
Oral and nasal cavities of mammals contain three distinct chemosensory epithelia.
1: Main olfactory epithelium containing sensory cells with odorant receptors in the nose.
2: Taste sensory epithelium of the taste buds on the tongue, soft palate, and epiglottis, a flap that is made of elastic cartilage tissue covered with a mucous membrane, attached to the entrance of the larynx. And,
3: The vomeronasal organ, an auxiliary olfactory sense organ found in many animals made up of a tubular structure in the nasal septum that contains sensory cells with pheromone receptors.
These chemosensory cells relay signals to the brain – more specifically to the cortex, the amygdale and hypothalamus. The chemosensory receptor can be grouped into three classes.
Each neuron of the main olfactory sensory system sends an axon, also known as a nerve fiber, a long and slender projection of a nerve cell, or neuron, that typically conducts electrical impulses away from the neuron’s cell body, to specific glomeruli, which is a small cluster of nerve fibers, and further to the main olfactory bulb and the olfactory nerve. Olfactory receptors are expressed in the cell membranes of olfactory receptor neurons and are responsible for the detection of odor molecules. The multigene family of olfactory receptors consists of over 900 genes in humans and 1500 genes in mice. Stimulated olfactory receptors activate a signal transduction cascade which produces a nerve impulse that is transmitted to the brain. Olfactory receptors are members of the class A rhodopsin-like family of G protein-coupled receptors. In addition, the bitter taste sensory system connects axonal projections of receptor cells in the taste sensory epithelium of the taste buds to gustatory nuclei in the brain stem. A subset of solitary microvillar cells are especially concentrated in the anterior part of the respiratory epithelium in the nasal cavity. These epithelial cells were originally characterized by the expression of T2R “bitter-taste” receptors, α-gustducin, and PLC(β2). TRPM5 is thought to serve as a downstream channel in this signal transduction pathway. These cells form synaptic-like contacts with trigeminal afferent nerve fibers, which carry the sensory information into the brain. Since irritating odorants at relatively high concentrations induce electrical signals in the anterior respiratory epithelium and cause Ca2+ elevation in dissociated TRPM5-positive cells it is thought that these solitary chemosensory cells are probably involved in sensing harmful or irritant chemicals that trigger protective reflexes such as sneezing and apnea and are mediated by the trigeminal system.
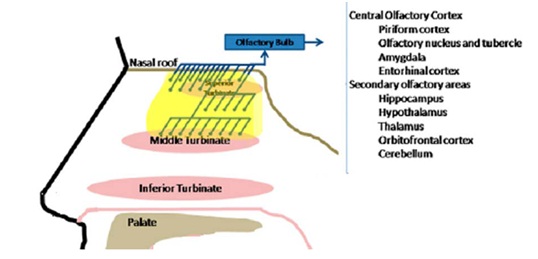
Neuroanatomic connections of the olfactory system. Shown is a sagittal cross-section of the lateral nasal wall. Olfactory neurons are depicted in blue, and their axons form filia of the olfactory nerve, which crosses the cribiform plate, synapse in the olfactory bulb, and continue to the various portions of the central nervous system. [Source: Patel and Pinto in Clinical Anatomy 27:54-60(2014). Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
-.-
Patel and Pinto in 2014 state “Olfaction remains a critical part of human behavior and emotional experience, and clinical implications of olfactory loss is continuing to emerge as its prevalence increases and effects on quality of life are becoming more apparent.”
To gain insight into the human olfactory receptor gene family Malnic et al. in 2004 analyzed the human genome database to define the full repertoire of olfactory receptor genes. The scientists analyzed the subfamily structure, the chromosome locations of genes encoding of members of each subfamily, and the subfamily composition of each chromosomal locus that contains intact olfactory receptor genes. In addition, information of mammalian olfactory receptor genes with known odorant specificities was used to explore potential relationships between odor detection and olfactory receptor subfamilies and gene loci. The study showed that humans have 636 olfactory receptor genes and identified 339 intact olfactory receptor genes and 297 olfactory receptor pseudogenes. The researchers showed that these genes are unevenly distributed among 51 different loci on 21 human chromosomes (see figure below). Furthermore, sequence comparisons showed that the human olfactory receptor family is composed of 172 subfamilies. In the last 25 years close to 490 research papers were published about olfactory genes.
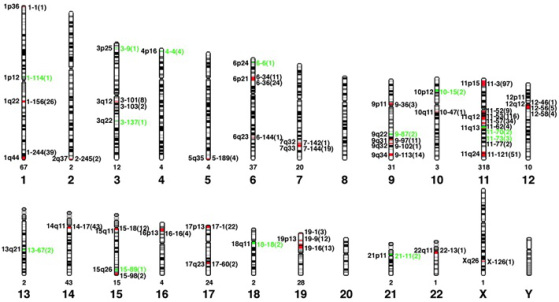
Chromosome locations of human OR genes as determined by Malnic et al. Six hundred thirty olfactory receptor genes were localized to 51 different chromosomal loci distributed over 21 human chromosomes. Olfactory receptor gene loci containing one or more intact olfactory receptor genes are indicated in red; loci containing only pseudogenes are indicated in green. The cytogenetic position of each locus is shown on the left, and its distance in megabases from the tip of the small arm of the chromosome is shown on the right. The number of olfactory receptor genes at each locus is indicated in parentheses, and the number of olfactory receptor genes on each chromosome is indicated below. Most human homologs of rodent olfactory receptors for n-aliphatic odorants are found at a single locus, chromosome 11p15. (Source: Malnic et al., 2004).
-.-
The subfamily assignments of the human olfactory receptor gene family for 339 human olfactory receptors can be found at: http://www.pnas.org/content/suppl/2004/02/03/0307882100.DC1/7882Table5.html.
-.-
Malnic et al. found that there is wide variation in the number of olfactory receptor genes at individual OR gene loci (1–116 OR genes) and on different chromosomes (0–318 OR genes). Furthermore, it was found that the percentage of olfactory receptor genes that are pseudogenes also varies among loci (0–100%). Of the 51 OR gene loci identified, 13 have only pseudogene(s), 38 have at least one intact OR gene, and 27 have more than one intact OR gene. Thus, 38 loci are potentially functional.
Potential associations between olfactory receptor gene loci and odorant recognition
Locus | OR (MOR#)* | Amino acid identity, % | Odorant(s) recognized | Perceived odor |
1q22 | OR23(267–13) | 87 | Lyral | Lemony, green |
5q35.3 | IG7(276–1) | 83 | Limonene | Lemon |
9q34.11 | ID3(136–6) | 71 | I-carvone | Spearmint, caraway |
11q12.1 | OR73(174–9)† | 82 | Eugenol | Spicy |
11q12.1 | OR74(174–4)† | 76 | Ethyl vanillin | Vanilla |
11q12.2 | OR912–93(175–1) | 66 | 2-Heptanone | Fruity |
11p15.4 | S25(204–32) | 74 | n-aliphatic alcohols | Herbal, woody, orange, rose |
11p15.4 | S46(32–4) | 69 | n-aliphatic acids | Rancid, sour, sweaty, fatty |
11p15.4 | S85(13–6) | 60 | n-aliphatic acids | As above |
11p15.4 | S86(8–2) | 67 | n-aliphatic acids | As above |
11p15.4 | S18(31–2) | 70 | n-aliphatic acids / alcohols | As above |
11p15.4 | S19(33–1) | 62 | n-aliphatic acids / alcohols | As above |
11p15.4 | S41(22–2) | 81 | n-aliphatic acids / alcohols | As above |
11p15.4 | S51(40–1) | 83 | n-aliphatic acids / alcohols | As above |
11p15.4 | S83(40–4) | 81 | n-aliphatic acids / alcohols | As above |
11p15.4 | I7(103–15) | 87 | n-aliphatic aldehydes | Fatty |
11q24.2 | M71(171–2) | 62 | Acetophenone | Floral / woody |
14q11.1 | S3(106–13P)† | 81 | n-aliphatic alcohols | Herbal, woody, orange, rose |
14q11.1 | S1(106–1)† | 87 | n-aliphatic acids | Rancid, sour, sweaty, fatty |
17p13.3 | hOR17–4 | 87 | Bourgeonal | Lily of the valley |
17p13.3 | hOR17–40 | 81 | Helional | Sweet, hay-like |
*Mouse OR designation according to ref. 7.; †OR73 and OR74 are in the same subfamily as are S1 and S3. Source: www.pnas.org_cgi_doi_10.1073_pnas.0401411101
-.-
More recently, Veerappa et al. in 2013 reported that the combinatorial effect of both ‘‘orthologous sequences obtained from closely related species’’ and ‘‘paralogous derived sequences’’ of human genotypic variations in the functional olfactory receptor repertoire provide the complexity of the occurring olfactory receptors in the central nervous system.
How did they do this?
The research group investigated 43 Indians along with 270 haplotype maps (HapMap) and 31 Tibetan samples to study genome variability and evolution. The analysis was performed using the Affymetrix Genome-Wide Human single nucleotide polymorphism (SNP) Array 6.0 chip, Affymterix CytoScan® High-Density array, high-density copy number variation (HD-CNV), and a “Multiple Alignment Fast Fourier Transform (MAFFT)” program. This allowed the researchers to observe a total of 1527 olfactory receptor genes in 503 CNV events from 81.3% of the study group, which included 67.6% duplications and 32.4% deletions encompassing more of genes than pseudogenes. Even though micro arrays do not provide as accurate data as next-generation-sequencing or direct sequencing does of the genes investigated this work is very impressive. Therefore, we now know that olfactory receptors belong to the largest family of genes and are highly polymorphic in nature having distinct polymorphisms associated with specific regions around the globe, and allow our species and mammals the detection of a large number of odor molecules with varying detection limits between species.
Sweet tasting molecules
What causes the different tastes of food? Many molecules, often also called chemical compounds, present in food sources have a sweet taste. Almost all simple carbohydrates or sugars taste sweet to some degree. Most people know how table sugar or sucrose tastes. Sucrose commonly known as table sugar or saccharose is a white, odorless, crystalline powder with a sweet taste, best known for its use in cakes, chocolate, ice cream and pastries. This disaccharide contains the monosaccharides glucose and fructose and has the molecular formula C12H22O11. It is the molecule we use or refer to when we want to create a sweet tasting dish. On the other hand, fructose, a fruit sugar, by itself tastes somewhat sweeter than sucrose. In addition, the taste of amino acids have long been described and the first amino acid isolated through gelatin acid hydrolysis was found to be so sweet that it was considered a sugar (‘‘sucre de gelatin’’). Amino acids are present in many foods and influence how delicious, sweet, bitter, good or bad many foods taste. Quantitative analysis of amino acids present in foods showed that the taste humans perceive largely depends on the kinds and amounts or ratios of amino acids found in these foods. The standard analysis to accurately quantify amino acids is the calssical “Amino Acid Analysis.” Furthermore, the perceived taste can range from sour or umami to sweet and bitter. Some of the amino acids also taste mildly sweet. Amino acids can occur in two forms, the L- (S-, or left) and D- (R- or right) form, which makes these molecules chiral or gives them “handiness.” In chemistry, chirality usually refers to molecules with structures that can be drawn as mirror images. Chiral molecules are also called enantiomers or optical isomers and pairs of enantiomers are designated as “right-” and “left-handed”. Therefore, the taste of different enantiomeric forms of amino acids has been investigated over the years because it represents the most impressive case of correlation between stereochemistry and flavor. In particular, Solms et al. in 1965 demonstrated that many L-amino acids are characterized by a sweet or bitter taste, while most D-amino acids primarily have a sweet flavor. Birch et al. in 1989 were able to demonstrate a relationship between the taste of enantiomeric amino acids and their physico-chemical properties, particularly the specific apparent volume, which represents the effective size of solutes in water due to their intrinsic molecular architecture. The results from this paper were used to interpret the steric exclusion of certain enantiomers from taste receptors. The amine acids alanine, glutamine, glycine, serine, threonine, and proline, taste sweet, whereas other amino acids are perceived as both sweet and bitter. Glutamate and aspartic acid taste sour. Umami is considered as one of the five basic tastes together with sweet, sour, bitter and salty. The Japanese word umami can be translated as savory taste. Glutamate receptors enable the umami taste in humans. The food additive monosodium glutamate or MSG was found to be responsible for the umami taste. Kikunae Ikeda in 1908 identified glutamate in the broth from kombu seaweeds as the molecule responsible for the umami flavor. A few years later, in 1913, the ribonucleotides inosine monophosphate (IMP) and guanosine monophosphate (GMP) were also identified to confer the umami taste. Furthermore, it was noticed that foods rich in glutamate and ribonucleotides taste more intense than foods that contain these ingredients alone. The amino acids arginine, histidine, isoleucine, leucine, methionine, phenylalanine, tryrosine, and valine were found to confer the bitter taste. In addition, many peptides and proteins can confer different types of taste. For example brazzein is a sweet-tasting protein extracted from the West African fruit of the climbing plant Oubli (Pentadiplandra brazzeana Baillon), and other examples of sweet tasting proteins are pentadin, monellin and thaumatin. On the other hand, in the sweet tasting stevia plant, steviol glycosides are responsible for the sweet taste of the plant (Stevia rebaudiana Bertoni) and the sweetness of some of these glycoside compounds can be 40 to 300 times sweeter than sucrose.
Selected references
Bassoli, A., Borgonovo, G., Caremoli, F., Mancuso, G.; The taste of D- and L-amino acids: In vitro binding assays with cloned human bitter (TAS2Rs) and sweet (TAS1R2/TAS1R3) receptors. Food Chemistry 150 (2014) 27–33.
Birch, G. G., & Kemp, S. E. (1989). Apparent specific volumes and tastes of amino acids. Chemical Senses, 14(2), 249–258.
Faus I, Sisniega H (2004). “Sweet-tasting Proteins”. In Hofrichter M, Steinbüchel A. Biopolymers: Polyamides and Complex Proteinaceous Materials II (8 ed.). Weinheim: Wiley-VCH. pp. 203–209. ISBN 3-527-30223-9.
Malnic B, Godfrey PA, Buck LB.; The human olfactory receptor gene family. Proc Natl Acad Sci U S A. 2004 Feb 24;101 (8):2584-9.
Anna Menini (Editor); The Neurobiology of Olfaction in Frontiers in Neuroscience. Boca Raton (FL): CRC Press; 2010. ISBN-13: 978-1-4200-7197-9.
Yoshinori Mine, PhD, Eunice Li-Chan, Bo Jiang (Editors); Bioactive Proteins and Peptides as Functional Foods and Nutraceuticals. 2010 Blackwell Publishing Ltd. And Institute of Food Technologists.
Nelson, G., Chandrashekar, J., Hoon, M. A., Feng, L., Zhao, G., Ryba, N. J., & Zuker, C. S. (2002). An amino-acid taste receptor. Nature, 416(6877), 199–202.
Riddhi M. Patel and Jayant M. Pinto; Olfaction: Anatomy, physiology, and disease. Clinical Anatomy 2004 (27, pages 54–60). Article first published online: 22 NOV 2013 | DOI: 10.1002/ca.22338
Schiffman, S. S., Sennewald, K., & Gagnon, J. (1981). Comparison of taste qualities and thresholds of D- and L-amino acids. Physiology & Behavior, 27(1), 51–59.
Schmidt, C. L. A. (1938). The chemistry of amino acids and proteins. Baltimore, MD: Thomas Books Ed. Springfield Ill.
Shallenberger, R. S. (1993). Taste chemistry (1st ed.). Glasgow: Blackie Academic & Professional Ed..
Shallenberger, R. S., Acree, T. E., & Lee, C. Y. (1969). Sweet taste of D- and L-sugars and amino-acids and the steric nature of their chemo-receptor site. Nature, 221(5180), 555–556.
Solms, J., Vuataz, L., & Egli, R. H. (1965). The taste of L- and D-amino acids. Experientia, 21(12), 692–694.
Temussi, P. A., Lelj, F., & Tancredi, T. (1978). Three-dimensional mapping of the sweet taste receptor site. Journal of Medicinal Chemistry, 21(11), 1154–1158.
Veerappa AM, Vishweswaraiah S, Lingaiah K, Murthy M, Manjegowda DS, Nayaka R, Ramachandra NB.; Unravelling the complexity of human olfactory receptor repertoire by copy number analysis across population using high resolution arrays. PLoS One. 2013 Jul 3;8(7):e66843. doi: 10.1371/journal.pone.0066843. Print 2013.
-.-
BSI offers custom services for oligonucleotide, peptide and protein based research.

