As of December 2013 90 solved structures were available in the PubMed structural database related to the CRISPR Cas System!
A few these models are shown below!
A PubMed search for “CRISPR Cas” showed that close to 90 solved structures related to this system were available by the end of December in 2013. The model of the CRISPR associated Cse3 protein, in complex with RNA published by Gesner et al. in 2011 is shown below.
Image may be NSFW.
Clik here to view.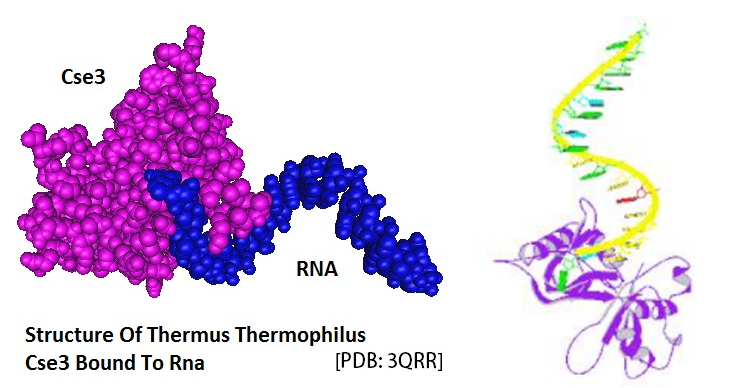
Figure 1: Model of the structure of Thermus Thermophilus Cse3 bound to RNA.
(Note: To generate the models PDB files were retrieved from the PubMed structure database and rendered using the modelling software packages Cn3D 4.3 and Pymol)
-.-
Jore et al. in 2011 reported the structure of CRISPR RNA (crRNA) which contains a 5’handle connected to a spacer sequence and a 3’handle. As can be seen from the figure below, the 3’handle is thought to form a hairpin structure. This type of loop is an unpaired loop of RNA that is created when an RNA strand folds and forms base pairs with another section of the same strand. The resulting structure is called a hairpin structure and looks like a loop or a U-shape.
Image may be NSFW.
Clik here to view.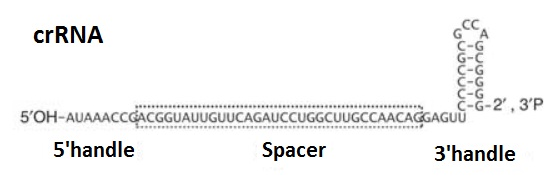
Figure 2: Structure of crRNA. (As proposed by Jore et al. in 2011).
Jinek et al. in 2012 reported models of the naturally occurring and engineered RNA-guided nuclease systems. The model for the naturally occurring RNA-guided nuclease system is shown below.
Image may be NSFW.
Clik here to view.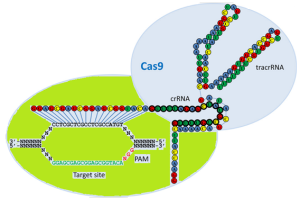
Figure 3: Schematic model of the naturally occurring RNA-guided nuclease systems.
The naturally occurring dual RNA-guided Cas9 nuclease is illustrated. crRNA interacts with the complementary strand of the DNA target site harboring an adjacent PAM sequence, shown as green and red text. TracrRNA base pairs with the crRNA, and the overall complex is recognized and cleaved by Cas9 nuclease shown in light blue color. Folding of the crRNA and tracrRNA molecules is predicted by the program Mfoldand the association of the crRNA to the tracrRNA is partially based on the model proposed by Jinek et al. (2012).
Spilman et al. in 2013 reported that they have solved the structure of an RNA Silencing Complex of the CRISPR Cas Immune System using cryoelectron microscopy. The research group reconstructed a functional Cmr complex bound with a target RNA at a resolution of approximately 12 A°. They showed that pairs of the Cmr4 and Cmr5 proteins form a helical core that is asymmetrically capped on each end by distinct pairs of the four remaining subunits: Cmr2 and Cmr3 at the conserved 50 crRNA tag sequence and Cmr1 and Cmr6 near the 30 end of the crRNA. The structure revealed that the shape and organization of the RNA targeting Cmr complex is strikingly similar to the DNA-targeting Cascade complex. In addition these results revealed a remarkably conserved architecture among very distantly related CRISPR Cas complexes.
The next figure shows the model of the functional Cmr complex bound to a target RNA.
Image may be NSFW.
Clik here to view.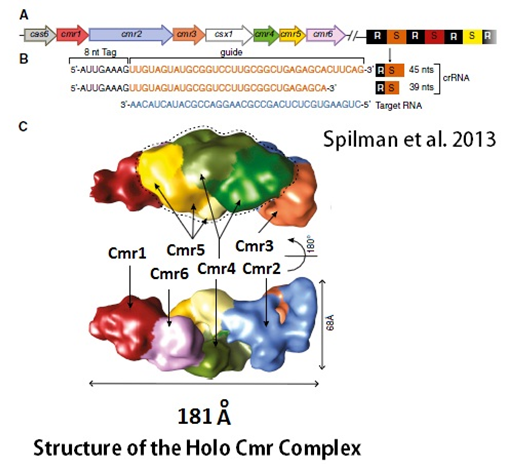
Figure 4: Overview of the P. furiosus CRISPR/Cas Locus and the Cmr Complex Structure
(adapted from Spilman et al. 2013). The structure was determined by cryoelectron microscopy.
(A) The genes encoding the Cmr complex protein subunits are color coded to match those used for structure images in subsequent figures. The CRISPR repeats are shown in black and spacers in various colors.
(B) The mature 39 nt and 45 nt crRNAs contain a 50 repeat-derived 8 nt sequence (50 tag) and a 31 nt or 37 nt spacer-derived sequence (guide), respectively. The sequence of the target RNA used in RNA cleavage assays and assembly with the Cmr complex is shown in blue.
(C) Color-coded EM density of the Cmr complex bound with the 45 nt crRNA and the target RNA is shown in two orientations. Cmr1, red; Cmr2, light blue; Cmr3, orange; Cmr4, three different shades of green; Cmr5, three different shades of yellow; and Cmr6, magenta.
Do you need long RNA oligonucleotides?
Modified or unmodified?
Biosynthesis Inc. can synthesize them for you!
Please inquire calling 1-800-227-0627
or by clicking www.biosyn.com
Image may be NSFW.
Clik here to view.
Reference
Gesner EM, Schellenberg MJ, Garside EL, George MM, Macmillan AM; Structure of Thermus Thermophilus Cse3 Bound to an RNA Representing a Product Complex. Nat.Struct.Mol.Biol. (2011) 18 p.688. PDB ID: 3QRR]
Matthijs M Jore, Magnus Lundgren, Esther van Duijn, Jelle B Bultema, Edze R Westra, Sakharam P Waghmare, Blake Wiedenheft, Ümit Pul, Reinhild Wurm, Rolf Wagner, Marieke R Beijer, Arjan Barendregt, Kaihong Zhou, Ambrosius P L Snijders, Mark J Dickman, Jennifer A Doudna, Egbert J Boekema, Albert J R Heck, John van der Oost & Stan J J Brouns; Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nature Structural & Molecular Biology 18, 529–536 (2011). doi:10.1038/nsmb.2019.
Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012; 337:816–821. [PubMed]
Michael Spilman,Alexis Cocozaki,Caryn Hale,Yaming Shao,Nancy Ramia,Rebeca Terns,Michael Terns,Hong Li,and Scott Stagg; Structure of an RNA Silencing Complex of the CRISPR-Cas Immune System. Molecular Cell 52, 146–152, October 10, 2013.