The CRISPR systems based gene editing method has been named the breakthrough technology of the year 2015 by the Journal Science. This gene editing method known as “clustered regularly interspaced short palindromic repeats, or short, CRISPRs”, is now ” ..poised to revolutionize research”, as was pointed out by Marcia McNutt, Editor-in-Chief, Science Journals. CRISPR systems are now heralded as the most promising revolutionary gene-editing methods useful for the discovery of new drugs or medicines.
CRISPR systems are now moving closer to therapeutic uses and raise high expectations for the discovery of new types of drugs and treatments. However, before these promises can become real and move on to human therapeutic uses, CRISPR systems need to be better understood. For example, more efficient and reliable tools will need to be developed. Specifically, CRISPR tools that allow selective, accurate and specific integration of gene fragments without off-target effects are needed. More control of the repair mechanism in the targeted cells, as well as quantitative delivery of CRISPR tools into the targeted cell, is also needed. Many research groups are now working to address these issues. To address these issues Kleinstiver et al. in early 2016 describe SpCas9-HF1, a high-fidelity CRISPR Cas9 nuclease variant harbouring alterations designed to reduce non-specific DNA contacts.
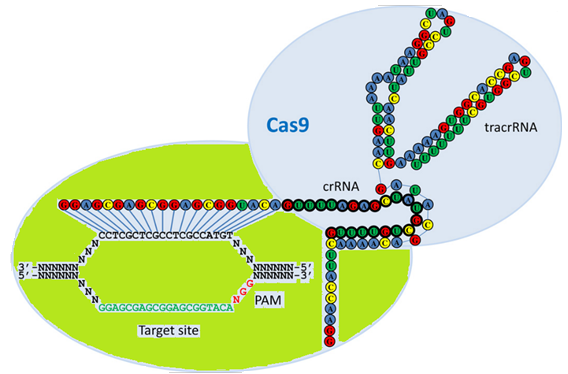
Figure 1: Schematic model of the naturally occurring RNA-guided nuclease systems.
This model of the naturally occurring dual RNA-guided Cas9 nuclease illustrates the interaction of crRNA with the complementary strand of the DNA target site harboring an adjacent PAM sequence, shown as green and red text. TracrRNA base pairs with the crRNA, and the overall complex is recognized and cleaved by Cas9 nuclease shown in light blue color. Folding of the crRNA and tracrRNA molecules was predicted by the program Mfold and the association of the crRNA to the tracrRNA is partially based on the model proposed by Jinek et al. (2012).
Potential applications for the CRISPR systems
- Cell engineering,
- Drug Discovery,
- CRISPR gRNA screening,
- siRNA screens,
- shRNA screens,
- Cell-based assays,
- Cell-line identification assays,
- Stem cell screening assays,
- CRISPR-based fine screening of the Human Genome,
- Epigenome editing,
- Libraries for next-generation sequencing (NGS),
- Determination of essential genes (House Keeping Genes),
- Development of better cell and animal models to test new drugs and therapies,
- New types of immuno-oncology treatments for cancer, Patient-derived animal models,
- Cancer and tumor screening and modeling,
- Transplantation models,
- Knockouts,
- Knockins,
- Tagging, Chromosomal rearagments,
- Transcriptional interference or activation,
- Genetic screening,
- And many more.
How do CRIPSRs work?
For CRISPR Cas9 to work, researchers can design a “guide RNA” (gRNA or crgRNA) to match the sequence of a specific target gene. The RNA guides the Cas9 enzyme to the desired target, where it then cuts the DNA. This cut inactivates the gene. Alternatively, CRISPR Cas9 allows replacing the sequence section adjacent to the cut with a different version of the gene. The CRISPR Cas9 system can be used in a variety of applications. Researchers now hope to use CRISPR Cas systems for the repair of defective genes that cause disease. To improve the system chemically modified RNA may be used in the usual guide RNA.
Every recently published paper covering advances made in gene editing methods based on CRISPRs suggest that this technology is a new game-changing technology. It appears that the CRISPRs are surpassing the zinc fingers and transcription activator-like effector nucleases (TALENS) as gene editing tools because of their ease-of-use and versatility. The use of the CRISPR technology offers the potential to pinpoint and repair genetic mutations, for example in the chimeric antigen receptor (CAR) T-cell field, by employing various gene editing strategies for the development of toolkits and therapies.
Presently, the CRISPR Cas system is considered to be the system of choice for gene editing. This gene-editing method now allows for a variety of promising applications useful as a genomic cut-and-paste method. Already the technique has been used to create “gene drives”, heralded as a method to eliminate pests or diseases they carry, and the first editing of the DNA of human embryos. This notion has prompted headlines and concerns and ethical considerations discussing possible human gene editing using CRISPRs.
The rapid pace of development in CRISPR-based gene editing methods has already started a debate if, how and when the technology should be used in human germline cells or embryos. On the positive site, this type of gene editing promises the elimination of devastating diseases. On the negative site, it is possible that tinkering with the genome crosses a moral boundary, specifically since all the mechanisms how the technique works are not well understood at this point in time. Also, the occurrence of possible off-target effects will need to be avoided for this gene editing method to work specifically.
The discovery of an unusual segment of neighboring DNA originally without any known biological function consisting of short repeating nucleotide sequences flanked by short unique segments let to a system, the bacterial CRISP Cas system, that now allows for easy manipulation of many genomes (Ishino et al. 1987). However, it took more than 20 years for scientists to figure out the significance of this system. Ultimately, the CRISPR Cas9 type II system that needs only one protein for it to function has become the system most used. The CRISPR Cas9 type II system uses a single endonuclease, Cas9. This enzyme together with guide RNA locates and cleaves invading DNA at specific sites containing conserved sequences called proto-spacer adjacent motifs (PAMs).
The CRISPR Cas systems allow detection of and protection against genetic elements. The type II system uses a single endonuclease, Cas9, that acts together with guide RNA to locate and cleave invading DNA at specific sites that are separated or distinguished from other sequence loci called proteo-spacer adjacent motifs (PAMs). The formation of the DNA targeting complex requires Cas9 and two distinct RNA transcripts, CRISPR RNA (crRNA) and trans-acting CRISPR RNA (tracrRNA).
Cleavage of specific DNA sequences
Researchers discovered that S. pyrogenes Cas9 nuclease, when used with a small CRISPR associated RNA (crRNA) and a separate transactivating RNA (tracerRNA), can be directed to cleave specific DNA sequences. (Jinek et al., 2012). The active complex contains a protein, Cas9 nuclease, and two RNA molecules, crRNA and tracrRNA.
Selective disruption of specific genes
Genes can be disrupted selectively as was shown by Sato et al., in 2015. The disruption of selected genes was achieved by combined injection of guideRNA and human Cas9 mRNAs. The research group reported the disruption of a gene (GGTA1) encoding the α-1,3-galactosyltransferase that synthesizes the α-Gal epitope using parthenogenetically activated porcine oocytes. They describe how it was done as follows: “After electric activation of in vitro-matured oocytes, these were cytoplasmically injected with a solution (~2 pL) containing in vitro synthesized hCas9 mRNA (2 ng/μL), gRNA (2 ng/μL; specific to GGTA1 exon 4), and EGFP mRNA (2 ng/μL) and were then cultured in vitro until blastocyst formation for approximately seven days.”
Repair of T cells
Schuman et al., also in 2015, reported that the delivery of Cas9 protein pre-assembled with guide RNAs enabled successful Cas9-mediated homology-directed repair in primary human T cells.
Targeted gene regulation
Qi et al., in 2013, showed that a catalytically dead Cas9 without endonuclease activity can be coexpressed with a guide RNA to generate a DNA recognition complex. This complex can specifically interfere with transcriptional elongation, RNA polymerase binding, or transcription factor binding. This system is called CRISPR interference (CRISPRi) and can efficiently repress expression of targeted genes in Escherichia coli. No off-target effects were detected. The CRISPRi system can be used to repress multiple target genes simultaneously in a reversible manner.
Mammalian cell engineering with Cas9 protein transfection
Liang et al, in 2015 described methods for the rapid synthesis of gRNA and the delivery of Cas9 protein/gRNA ribonucleoprotein complexes (Cas9 RNPs) into a variety of mammalian cells. Delivery into cells was achieved through liposome-mediated transfection or electroporation. The delivery of Cas9 and synthesis of guide RNA (gRNA) are steps that can limit the overall efficiency and ease of use of CRISPR Cas systems. The researchers reported nuclease-mediated indel rates of up to 94% in Jurkat T cells and 87% in induced pluripotent stem cells (iPSC) for a single target. An “indel” refers to the insertion or deletion of bases in DNA sequences. Key features of these methods are the turn-around time. The design to analysis can be done within 3 days using in vitro transcribed gRNA and cas9 protein or mRNA. Effective transfection of Cas9 mRNA or RNPs is achieved using Lipofectamine® 3000 or RNAiMax. Electroporation of cas9 RNPs is done in difficult cell lines (Jurkat, iPSC, CD34+). Cas9 RNPs enables multiple and simultaneous targeting of loci. These methods are reported to allow for high throughput set up and transfection in multi-well plates.
RNA library preparation
RNA library preparation protocols require small RNAs with 5’P and 3’OH groups. Therefore when using CRISPR Cas based systems for library constructions the right choice of protocol is crucial since certain subtypes of CRISPR Cas produce crRNAs with different chemical groups at their 5’ and 3’ ends. Pre-crRNA cleavage products generated by Cas6 endoribonucleases, such as Cas6c, Casgf (type I) and Cas6 (type III) contain 5’ hydroxyl and 2-3’ cyclic phosphate ends. These crRNA pools are not accessible for 5’-linker ligation and cannot be extended by poly(A) polymerase. A treatment with poly nucleotide kinase (PNK) is needed prior to the cDNA preparation step for capture in RNA-sequencing libraries.
Example of a cell engineering workflow
(Liang et al., 2015):
Day 1 | CRISPR Targets: Preparation involves the design of CRISPR targets, preparation or ordering of primers, and cell seeding. |
Day 2 | gRNA: Make gRNA. Assemble gRNA template. Perform IVT reaction and purify gRNA. A synthetic gRNA free of truncation and interfering byproducts can be used. Delivery: Deliver gRNA and Cas9 to cells. Incubate cells with gRNA-Cas9 protein complex with lipid based delivery or electroporation for 24 to 48 hours. |
Day 3 to 4 | Analyze results: Analyze editing efficiency by PAGE, PCR and/or sequencing. |
Parts of the CRISPR Cas system - Abbreviations
Below is a list of tools or parts the CRISPR systems need to function. Most of them can now be generated synthetically or via cloning of the various nucleases.
CRISPR Cas Tools or Parts | Short Description | Synthetic |
Cas | CRISPR-associated genes are located in the vicinity of CRISPR array and are necessary for the silencing of invading nucleic acid. | |
Cas9 nuclease | Cas9 (CRISPR associated protein 9). A RNA-guided DNA endonuclease enzyme associated with the CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) adaptive immunity system in Streptococcus pyogenes and other bacteria. The Cas9 protein in its active form can modify DNA utilizing crRNA as its guide. Many variants exist with differing functions such as single strand nicking, double strand break, or DNA binding due to Cas9's DNA site recognition function. | mRNA encoding different Cas nucelases. Can be injected into cells. hCas9 mRNA. |
Cas9t | Cas9–crRNA–tracrRNA ternary complex, which functions as an RNA guided DNA endonuclease and mediates site-specific DNA cleavage. | |
Clustered regularly interspaced short palindromic repeat (CRISPR) | An array of short conserved repeat sequences interspaced by unique DNA sequences of similar size called spacers. They often originate from phage or plasmid DNA. CRISPR array together with Cas genes form the CRISPR Cas system, which functions as an adaptive immune system in prokaryotes. | |
CRISPR RNA (crRNA) | Small RNA molecule generated by transcription and processing of the CRISPR array. crRNA is composed of a conserved repeat fragment(s) and a variable spacer sequence, which matches the complimentary sequence in the invading nucleic acid. Contains a target-specific protospacer of 20 to 40 bases and a repeat motif that is recognized and bound by the tracrRNA. Recognition and binding is usually occurring at a hairpin loop forming the active complex with Cas9. | |
gRNA or guide RNA | During gene editing, guideRNAs (gRNAs) can be synthesized to perform the function of the tracrRNA:crRNA complex. Recognizing gene sequences need to have a PAM sequence at the 3'-end. gRNAs allow transportation of Cas9 to anywhere in the genome during gene editing. However, no editing can occur at any site other than the one at which Cas9 recognizes PAM. | |
Homologous repair (HR) | Error-free DNA repair pathway that seals the broken DNA molecule using a homologous sequence (template). | |
Indel | The term indel refers to the insertion or the deletion of bases in DNA sequences of an organism. | |
Non-homologous end joining (NHEJ) | A pathway that repairs DNA double strand breaks (DSB) in the absence of a homologous template; usually leads to small insertions or deletions. | |
PAM or protospacer adjacent motif | A short conserved nucleotide stretch located in the vicinity of a protospacer in the target DNA and necessary for DNA cleavage by Cas9t. A DNA sequence immediately following the DNA sequence targeted by the Cas9 nuclease. PAM is part of the invading virus or plasmid DNA but not a component of the bacterial CRISPR locus. If the target DNA sequence is not followed by the PAM sequence Cas( will not be able to successfully bind to and cleave the target DNA. | |
Protospacer | A fragment in the target DNA, which matches a spacer sequence in the CRISPR array. | |
sgRNA | Single guide RNAs are a combined RNA consisting of a tracrRNA and at least one crRNA. RNA hairpin obtained by connecting crRNA and tracrRNA into a single molecule. | Synthetic sgRNA can be used for CRISPR experiments if they |
tracrRNA or trans-activating crRNA | A small trans-encoded RNA. TracrRNA is a complementary RNA sequence to and base pairs with a pre-crRNA forming an RNA duplex. This duplex is cleaved by RNase III, an RNA-specific ribonuclease, forming a crRNA/tracrRNA hybrid. This hybrid acts as a guide for the endonuclease Cas9 that cleaves the invading nucleic acid. | |
Transcription activator-like effector nuclease (TALEN) | An artificial nuclease obtained by fusing Xanthomonas transcription activator-like effector (TALE) DNA binding domains to the nonspecific nuclease domain. | |
Trans-acting CRISPR RNA (tracrRNA) | Trans-encoded small RNA molecule that forms a duplex with a repeat fragment of crRNA. | |
Triple helix forming oligonucleotide (TFO) | An artificial oligodeoxynucleotide, which binds to the polypurine sequences of the double-stranded DNA forming DNA triple helix. | |
Zinc finger nuclease (ZFN) | An artificial nuclease created by fusing zinc finger motifs, which serve as DNA recognition modules, to a nonspecific DNA cleavage domain of the FokI restriction endonuclease. |
Specific Primers and adaptors useful for CRISPR experiments
The CRISPR systems can be studied using RNA-sequencing techniques. Several studies have already used RNA-sequencing for crRNA analysis. Using this approach, primary and processed transcripts can be distinguished and transcriptional start sites can be annotated fro whole genomes. Also, CRISPR can be used for the typing of Myobacterium tuberculosis complex (MTC) and Salmonella enteric serotypes. During MTC spoligotyping a single couple of primers (DRa-DRb) allows amplification of a set of overlapping fragments that can be detected by hybridization by PCR. For the detection, one primer is biotinylated and a detectable reporter is added. Usually streptavidin-peroxydase using chemoluminescence detection or streptavidin-phytoerythrin using microbead-base laser detection is use for this approach. There is no end in sight for new types of CRISPR based applications. Below is a list of oligonucleotide sequences as examples for CRISPR based applications.
Primers or adaptors | Sequence | Use & Notes |
cDNA Library Preparation | Heidrich et al. 2013 | |
Repair template | DNA that guides the cellular repair process allowing insertion of a specific DNA sequence. | Insertion of DNA sequence. |
RNA adaptor | 5’-UUU CCC UAC ACG ACG CUC UUC CGA UCU-3’ | 5’ Illumina sequence adapter ligated to the 5’-phosphate of RNAs. |
Oligo(dT)-adapter primer | 5’-GTG ACT GGA GTT CAG ACG TGT GCT CTT CCG ATC TTT TTT TTT TTT TTT TTT TTT TTT TVN-3’ | 3’ Illumina sequencing adapter primer used for cDNA synthesis together with M-MLV reverse transcriptase. |
Tru-Seq-Sense_Primer | 5’-AAT GAT ACG GCG ACC ACC GAG ATC TAC ACT CTT TCC CTA CAC GAC GCT CTT CCG ATC T-3’ | Adapter sequence flanking cDNA inserts. |
Tru-Seq-Antisensce_Primer plus 6-mer barcode | 5’-CAA GCA GAA GAC GGC ATA CGA GAT NNN NNN GTG ACT GGA GTT CAG ACG TGT GCT CTT CCG ATC (dT25)-3’ | Adapter sequence flanking cDNA inserts. |
CRISPR loci (CRISPR2 repeat sequence) | 5-CGG TTT ATC CCC GCT GGC GCG GGG AAC AC-3’ | 29 bp in Salmonella |
| MTC-Spoligotyping | |
DRa | 5’-GGT TTT GGG TCT GAC GAC-3’ | MTC-Spoligotyping |
DRb | 5’-CCG AGA GGG GAC GGA AAC-3’ | MTC-Spoligotyping |
DRSTMA | 5’-CCG CTG GCG CGG GGA ACA-3’ | STM-CRISPOL |
| Priming | |
Priming Protospacer pg8_F | 5’-ATGTTG TCT TTC GCT GCT GAG GGT GAC GAT CCC GC-3’ | Plasmid generation. |
Priming Protospacer pg8_R | 5’-GCGGGATCG TCA CCC TCA GCA GCG AAA GAC AAC AR-3’ | Plasmid generation. |
| Substitutions at the seed region (CIT) are highlighted in bold. And the functional ATG PAM sequences are underlined. | |
Oligonucleotide T4B_7-F | 5’-TTT TTGGATCCG CGA CTT TAC CAG CGA STG-3’, BamHI restriction site | T4 phage insertion |
Oligonucleotide T4B_7-R | 5’-TT TTGAGCTCG GTA ATG CAG CTT CAG GAA AA-3’, ,SacI restriction site | T4 phage insertion |
| CRISPR I repeat spacer | |
g8-repeat | 5’-CTG TCT TTC GCT GCT GAG GGT GAC GAT CCC GC-3’ | |
| CRISPR expansion check sequence | |
Promoter-g8 spacer Ec_LDR-F | 5’-AAG GTT GGT GGG TTG TTT TTA TGG-3’ | |
Promoter-g8 spacer M13_g8 | 5’-GGA TCG TCA CCC TCA GCA GCG-3’ | |
Molecular marker data | ||
http://www.pasteur-guadeloupe.fr:8081/SITVITDemo | ||
5’ - Protospacer Sequences plus PAM -3’ | ||
CCA TAC CAA ACG ACG AGC GTG ACA CCA CGA TGA AG | pT7blue nP vector | |
{CRISPR, Methods and Protocols. 2015 MMB 1311} | TAT ATA TGA GTA AAC TTG GTC TGA CAG TTA CCA AG | pT7blue P vector |
TTG GCC GCA GTG TTA TCA CTC ATG GTT ATG GCA AG | pT7blue P vector | |
TCA TTC TGA GAA TAG TGT ATG CGG CGA CCG AGA AG | pT7blue P vector | |
TGC TCA TCA TTG GAA AAC GTT CTT CGG GGC GAA AG | pT7blue P vector | |
AAA GAA GAC GTA TTC AAC CCG GAT ATG CGA ATA AG | T4 P | |
ACC CGA CTA GAT GGG GAT ATG AAG ATA ATC TCA AG | T4 P | |
GAA CCA CGA TAT ATT CAT TCG TGC ATC TAT TTA AG | T4 P | |
ATG CTA TTG AAC ACA TTC CGG TAT CAG GAA CAA AG | T4 P | |
CAA ATC CTT TCC TTT AAC CCC ACG AAT AAT TTA AG | T4 P | |
ATA ACA CTT GAA TCA TTC ATC TAT TTT AAC CTT AG | T4 P |
Note 1: Results from clones containing CRISPR cassettes expanded by a single repeat-spacer unit. . The sequence including the PAM and the source of the spacer is listed. (For example, T4 insert of the pT4acq plasmid). The location of the PAM is highlighted in red. CRISPR cassettes expanded by single repeat unit. CRISPR cassettes indicate dynamics of CRISPR-phage interactions in metagenomes.
Note 2: HLA-DR: HLA-DR is an MHC class II cell surface receptor. This receptor class is encoded by the human leukocyte antigen complex on chromosome 6 region 6p21.31. The complex of HLA-DR (Human Leukocyte Antigen - antigen DRelated) with its ligand is a ligand for the T-cell receptor (TCR). The HLA-DR ligands are peptides of 9 amino acids in length or longer.
Note 3: CRISPR cassettes are transcriped as long pre-crRNAs and are processed into short crRNAs by one of the Cas proteins. crRNAs contain variable central sequences corresponding to CRISPR spacer which are flanked by fragments of CRISPR repeat sequences. crRNAs are bound by Cascade. Cascade is a complex of several Cas proteins. The Cascade-crRNA complex recognizes dsDNA containing a protospacer matching the crRNA spacer. The presence of the PAM sequence increases the strength of the interactions in vitro. Artificial nucleic acdis, such as bridged nucleic acids (BNAs), can be inserted into the PAM region of synthetic crRNAs to increase the binding strength of the crRNA. When the target is recognized an R-loop containing an extended RNA-DNA heteroduplex is formed. The heteroduplex involves the entire length of the spacer-protospacer. Successful target recognition leads to target cleavage.
Reference
Brouns SJ (2012). "A swiss army knife of immunity.". Science 337 (6096): 808–9. doi:10.1126/science.1227253. PMID 22904002.
Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, et al. (2011). "CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III.". Nature 471 (7340): 602–7. doi:10.1038/nature09886. PMC 3070239. PMID 21455174.
Gogleva, A. A., Gelfand, M. S., & Artamonova, I. I. (2014). Comparative analysis of CRISPR cassettes from the human gut metagenomic contigs. BMC Genomics, 15(1), 202. http://doi.org/10.1186/1471-2164-15-202
Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A (December 1987). "Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product". Journal of Bacteriology169 (12): 5429–5433. PMC 213968. PMID 3316184.
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E.; A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012 Aug 17;337(6096):816-21. doi: 10.1126/science.1225829. Epub 2012 Jun 28.
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012). "A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity.". Science 337 (6096): 816–21. doi:10.1126/science.1225829. PMID 22745249.
Kleinstiver, Benjamin P., Pattanayak, Vikram, Prew, Michelle S., Tsai, Shengdar Q., Nguyen, Nhu T., Zheng, Zongli, Keith Joung, J.; High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature 2016/01/06/online, advance online publication. 1476-4687. http://dx.doi.org/10.1038/nature16526., 10.1038/nature16526.
Le Rhun A, Charpentier E (2012). "Small RNAs in streptococci.". RNA Biol 9 (4): 414–26. doi:10.4161/rna.20104. PMID 22546939.
Xiquan Liang,Jason Potter, Shantanu Kumar, Yanfei Zou, Rene Quintanilla, Mahalakshmi Sridharan, Jason Carte, Wen Chen, Natasha Roark, Sridhar Ranganathan, Namritha Ravinder, Jonathan D. Chesnut; Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. Journal of BiotechnologyVolume 208, 20 August 2015, Pages 44–53. http://www.sciencedirect.com/science/article/pii/S016816561500200X
Lundgen, Charpentier, Fineran (ed.); CRISPR, Methods and Protocols. 2015 Methods in Molecular Biology 1311. Springer Series. Humana Press.
Marraffini, L. A., & Sontheimer, E. J. (2010). CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nature Reviews. Genetics, 11(3), 181–190. http://doi.org/10.1038/nrg2749.
Masahiro Sato, Miyu Koriyama, Satoshi Watanabe, Masato Ohtsuka, Takayuki Sakurai, Emi Inada, Issei Saitoh, Shingo Nakamura and Kazuchika Miyoshi; Direct Injection of CRISPR/Cas9-Related mRNA into Cytoplasm of Parthenogenetically Activated Porcine Oocytes Causes Frequent Mosaicism for Indel Mutations. Int. J. Mol. Sci. 2015, 16, 17838-17856; doi:10.3390/ijms160817838
Pougach, K., Semenova, E., Bogdanova, E., Datsenko, K. A., Djordjevic, M., Wanner, B. L., & Severinov, K. (2010). Transcription, Processing, and Function of CRISPR Cassettes in Escherichia coli. Molecular Microbiology, 77(6), 1367–1379. http://doi.org/10.1111/j.1365-2958.2010.07265.x
Ekaterina Savitskaya, Ekaterina Semenova, Vladimir Dedkov, Anastasia Metlitskaya & Konstantin Severinov (2013) High-throughput analysis of type I-E CRISPR/Cas spacer acquisition in E. coli, RNA Biology, 10:5, 716-725, DOI: 10.4161/rna.24325. http://dx.doi.org/10.4161/rna.24325.
Kathrin Schumanna, Steven Linc, Eric Boyera, Dimitre R. Simeonova, Meena Subramaniame,f, Rachel E. Gatee,f, Genevieve E. Haliburtona,b, Chun J. Yee , Jeffrey A. Bluestonea , Jennifer A. Doudnac, and Alexander Marsona; Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. PNAS 2015, 112, 33, 10437-10442. http://www.pnas.org/content/112/33/10437
Terns MP, Terns RM (2011). "CRISPR-based adaptive immune systems.". Curr Opin Microbiol 14 (3): 321–7. doi:10.1016/j.mib.2011.03.005. PMC 3119747. PMID 21531607.
UC San Diego Health https://health.ucsd.edu/news/releases/Pages/2015-11-16-RNA-Based-Drugs-Give-More-Control-Over-Gene-Editing.aspx
.jpg)
.png)