In summary, antiviral peptides may offer hope for the design of better and less expensive therapeutics to help prevent viral infections.
Reference
The Chemical Synthesis of OligonucleotidesThe study of nucleic acids has now become a fruitful and dynamic scientific enterprise. Nucleic acids are of unique importance in biological systems because genes are the fundamental unit of heredity. The process how genes are expressed in all living organisms is fundamentally important and most genes are located in the chromosomes within the cell nucleus. Genes express themselves via a protein machinery in the cytoplasm. The genetic material was identified as deoxyribonucleic acid (DNA) in 1944 (Avery et al. 1944). Next, the double-helical nature of DNA was revealed in 1953 by Francis Crick, James Watson and Maurice Wilkins. Furthermore, the combined action of multiple genes defines the properties or phenotype of higher organisms. Even to this date multi-gene characteristics are difficult to analyze. Genes are made up of deoxyribonucleic acid or DNA, and each gene is a linear segment, or polymer, of a long DNA molecule. However, genes have been found to be nonrandomly distributed on the chromosomes and vary enormously in size and intron-exon structure. A DNA polymer, or DNA oligonucleotide, contains a linear arrangement of subunits called nucleotides. There are four types of nucleotides. Each nucleotide has three components; a phosphate group, a sugar and a base that contains nitrogen within its structure. The sugar moiety in DNA oligonucleotides is always dexoyribose, and there are four alternative bases: adenine (Ade, A), thymine (Thy, T), guanine (Gua, G), and cytosine (Cyt, C). The phosphate groups and the deoxyribose sugars form the backbone of each DNA stand. The bases are joined to the deoxyribose sugar and stick out to the side. Both, DNA and ribonucleic acid (RNA), consist of 5’-3’ phosphodiester-linked nucleotide units that are composed of a 2’-deoxy-D-ribose (DNA) or D-ribose (RNA) in their furanose forms and a heteroaromatic nucleobase (A, T, G, and C; A, U, G, C), and the resulting oligonucleotide chain is composed of a polar, negatively charged sugar-phosphate backbone and an array of hydrophobic nucleobases. The amphiphilic nature of these polymers dictates the assembly and maintenance of secondary and tertiary structures the oligonucleotides can form. In double stranded DNA, the bases of one strand are paired with the bases in the other strand. Adenine (A) in one strand is paired with thymine (T) in the other strand and guanine (G) in one strand is paired with cytosine (C) in the other strand as well. In the DNA duplex structure, genetic information is stored as a linear nucleotide code. This code can be accessed and replicated. RNA, or ribonucleic acid, is another structurally related essential biopolymer. RNA differs from DNA in having the sugar ribose in place of the deoxyribose. Furthermore, in RNA the thymine (T) is replaced with uracil (U). The bases A and G are purine bases as they contain a double ring structure called the purine ring. The other two bases, C and T, are pyrimidine bases because they contain a single pyrimidine ring. Hydrogen bonds hold the two strands together.
Figure 1: Structures of nucleic acids. Figure 2: Atomic numbering and definitions of torsion angles for one nucleotide according to the UPAC nomenclature. The designation of chain direction and main chain atoms of i th unit in a polynucleotide chain and the atom numbering for the bases of common nucleosides and nucleotides are illustrated. Hydrogen atoms carry the same numbers as the heavy atoms to which they are attached. The name in parenthesis applies when the 'd' in parenthesis in the formula is present. (Source: www.chem.qmul.ac.uk/iupac/misc/pnuc1.html).
Figure 3: Representative types of base pairs. Purines and pyrimidines can form base pairs through hydrogen bonds. Watson-Crick, Hoogsten and wobble base pairs are the most common. An extensive list of graphical base pairs can be found in “Wolfram Sanger: Principles of Nucleic-Acid Structure. Spinger-Verlag New York Berlin Heidelberg Tokyo, pp 120.” In biological systems, nucleic acids exist in higher ordered structures held together through self-assembly. Since hydrogen bonding and base stacking are the major driving force for this self-assembly the structural properties of nucleotides can affect the self-assembly process. These molecular forces dictate the final tertiary structure of nucleic acids (The RNA World). The central dogma of molecular biology, originally described by Francis Crick, illustrates how DNA is transcribed into RNA which is ultimately translated into protein sequences. According to the dogma nucleic acid alone may store information or specify the sequence of gene products, proteins. Proteins are never able to specify a nucleic acid or protein sequence.  Figure 4: The central dogma of molecular biology.  Figure 5: Structures of oligonucleotides. In the beginning years, the chemical synthesis of specific oligoribo- and oligodeoxyribonuclotides has been regarded as a somewhat esoteric chemistry of natural compounds. The advent of gene technology and the development of new chemical and analytical methods such as high performance liquid chromatography (HPLC), 31P-NMR spectroscopy and automated synthesis methods has changed this all. The development and introduction of commercially available “gene synthesizing machines” following the development of chemical DNA and RNA synthesis strategies has made the synthesis of natural and artificial oligo-nucleotides a routine procedure as well as more cost-efficient and faster. To allow for a successful oligonucleotide synthesis the following fundamental prerequisites need to be established:
Several strategies have been developed in the past which comply with these limiting provisions. The phosphodiester and the phosphotriester approaches, both, utilize protection of the 3’ and 5’ hydroxyl groups of the deoxyribose. The phosphotriester uses a third protecting group for the protection of the internucleotide bond. A third approach called the “phosphate” procedure employs compounds with trivalent phosphorus and can be regarded as a trimester method. The development of oligonucleotide synthesis started with the phosophotriester method in 1955 but the fist significant success is reported to be achieved using the phosphodiester method when the genes for alanine and tyrosine suppressor tRNAs of yeast and E. coli were synthesized in the 1980s. Figure 6: Outline of the synthesis of the dinucleotide d(TpT) by Michelson and Todd reported in 1955. In this approach, 3’-O-acetylthymidine was phosphorylated with a phosphorochloridate, and the phosphate group was protected with a benzyl group. The structure of the dinucleotide was confirmed by enzymatic digestion (Michelson & Todd, 1955). This synthesis approach later became known as the phosphotriester approach. The literature reports that Gobind Khorana, in 1956, accidentally discovered the phosphodiester method for the chemical synthesis of deoxyribo-oligonucleotides (Khorana et al., 1956). The exploitation and further development of this method by many scientists in subsequent years for the chemical synthesis of deoxyribo-oligonucleotides led to the elucitation of the genetic code and the first total synthesis of a gene (Khorana et al., 1956; Khorana, 1961, 1969, 1979). Marshall Nirenberg and Gobind Khorana broke the genetic code and could assign code words called codons. Codons are triplets of nucleotides coding for the twenty amino acids. Both scientists received the Nobel Prize in Physiology or Medicine in 1968 together with Robert Holley. Phosphotriester Method Where B = A,C, G or T nucleo bases; R1, R2, R3 = protecting groups; X = Halogen Figure 7: General outline of the phosphotriester method. Figure 8: Outline of Khorana’s synthesis approach later known as the phosphodiester approach. In this approach, 5’-O-tritylthymidine and 3’-O-acetylthymidine 5’-phosphate are reacted in the presence of toluene-4-sulfonyl chloride (TsCl) or N1,N3-dicyclohexylcarbodiimide (DCC) in a pyridine solution. The removal of the trityl and acetyl group yields the d(TpT) dinucleotide (Khorana et al., 1956; 1957; Gilman and Khorana, 1958). Posphodiester Method Where B = A,C, G or T nucleo bases; R1, R2 = protecting groups Figure 9: General outline of the phosphodiester method.In the phosphodiester method the phosphate group between the two nucleotides is unprotected which makes the resulting compounds only soluble in organic solvents to a limited extent. Phosphite Method Where B = A,C, G or T nucleo bases; R1, R2, R3 = protecting groups; X = Halogen, N(CH3)2, morpholine Figure 10: Chemistry of phosphite method.Willi Bannwarth in 1985 reported a simple synthesis of phosphoramidite dinucleotides with two different phosphorous-protecting groups for the synthesis of 2′-oligodeoxynucleotides on a polymer support called the “Phosphite-Triester Method. Even though oligonucleotides can be assembled manually in a step-wise fashion this process has been and is quite laborious and demanding. The availability of commercial DNA synthesizers has made the process easier and more cost effective. The principle of solid phase synthesis was first developed and applied to the synthesis of polypeptides by Robert Bruce Merrifield (July 15, 1921 – May 14, 2006), an American biochemist who won the Nobel Prize in Chemistry in 1984 for the invention of solid phase peptide synthesis. He realized that the key to a successful synthesis is to anchor the first monomer to an insoluble polymeric support. Other monomers can then be joined, one by one, to the fixed terminal end of the growing polymer. At the end of the synthesis, the completed polymer chain can be detached from the insoluble polymer and purified. This process has been further optimized over the years to become highly efficient and has now become a fundamentally important method employed in automated oligonucleotide synthesizers. I. General Methods of Solid Phase Oligonucleotide Synthesis.a) Phosphoramidite method The phosphoramidite method of DNA synthesis is currently considered as the standard synthesis method used in most automated synthesizers today. This method allows achieving the high coupling efficiencies needed to synthesize longer and longer oligonucleotides with low amounts of failure sequences. The oligonucleotide phosphoramidite synthesis chemistry was introduced nearly 20 years ago (McBride and Caruthers, 1983). Building blocks used for synthesis are commonly referred to as “monomers”, which are activated DNA nucleosides (phosphoramidites). The dimethoxytrityl (DMT) group is used to protect the 5’-end of the nucleoside, a β-cyanoethyl group protects the 3’-phosphite moiety, and may also include additional groups that serve to protect reactive primary amines in the heterocyclic nucleo bases. The protecting groups are selected to prevent branching or other undesirable side reactions from occurring during synthesis. Oligonucleotides are synthesized on solid supports. Typically, the support is a small column filled with control pore glass (CPG), polystyrene or a membrane. The oligonucleotide is usually synthesized from the 3’ to the 5’. The synthesis begins with the addition of a reaction column loaded with the initial support-bound protected nucleotide into the column holder of the synthesizer. The first nucleotide building block or monomer is usually anchored to a long chain alkylamine-controlled pore glass (LCAA-CPG). A schematic diagram general outline the solid phase oligonucleotide synthesis of a dinucleotide is illustrated below.  The phosphoramidite approach to oligonucleotide synthesis proceeds in four (4) steps, which are schematically outlined in figure 11. Automated synthesis is done on solid support, usually controlled pore glass (CPG) or polystyrene. Synthesis is initiated with cleavage of the 5’-trityl group by brief treatment with dichloroacetic acid (DCA) dissolved in dichloromethane (DCM). Next, the monomer activated with tetrazole is coupled to the available 5’-hydroxyl resulting in a phosphite linkage. Subsequent phosphite oxidation by treatment with iodine using a THF/pyridine /H2O solution yields a phosphate backbone. The capping step with acetic anhydride, which terminates undesired failure sequences, completes the cycle of oligonucleotide synthesis. As figure 12 illustrates a typical synthesis cycle includes a condensation, a capping, an oxidation, and a cleavage or deprotection step. In general, automated DNA oligonucleotide synthesis produces a single-stranded oligonucleotide product per column.  After the final sequence has been assembled, the oligomer must be removed by cleaving it from the support and fully deprotected prior to its use. A 90 minute treatment with ammonium hydroxide at room temperature can be used to cleave the oligomer from the support and to deprotect the phosphorous by β–elimination of the cyanoethyl group. The acetyl capping groups and the base protecting groups are more difficult to remove and a 24 hour treatment at room temperature or an overnight treatment at 55 °C with ammonium hydroxide allows for effective removal of these groups. After cleavage and deprotection, the resulting crude mixture contains the product oligomer, possible truncated failure sequences with free 5’hydroxyl ends, byproducts of deprotection, and silicates from hydrolysis of the glass support. Different purification methods can be used to separate the product oligonucleotide from the contaminating species. b) H-Phosphonate method. The first internucleotide reactions employing H-phosphonates were reported in the 1950s (Hall et al., 1957), and the first solid supported oligonucleotide synthesis was described in the early 1970s (Kabachnik et al., 1971), where the reported coupling efficiency was between 46 and 80%. The modern H-phosphonate method involves the use of triethylamine (TEA) or 1,8-Diazabicyclo[5.4.0]undec-7-ene salts, or DBU salts of corresponding 3’-H-phosphonate monomers, which need to be activated with the appropriate acetyl chloride followed by coupling of activated O-acetyl phosphonate to the 5’-hydroxyl of the previous nucleoside attached to solid support (Figure 13). Isopropyl-H-phosphonate together with the same activator is used for the capping step of the cycle.  The resulting poly-H-phosphonate backbone can be oxidized after the final oligonucleotide is assembled from solid support in one step. The coupling efficiency of H-phosphonate synthesis is usually 94-95%, which is quite low compared to the phosphoramidite method. This approach is mainly used in cases when the desired modification of the phosphorous backbone can be done only through an H-phosphonate intermediate such as by using boranophosphates (Sergueev & Shaw, 1998) or phosphoramidates (Froehler, 1986), or others. c) Phosphotriester method. The phosphotriester approach (Letsinger & Mahadevan, 1965, 1966) is another successful method of oligonucleotide synthesis on solid support. Usually the coupling phosphorylating agent is an activated HOBt phosphate ester (Figure 14).  II. RNA synthesis.Automated RNA synthesis on solid support was introduced a decade later than DNA synthesis and many disappointing results had to be overcome leading to a difficult history of development. The use of one extra 2’-hydroxy group makes this chemistry magnitudes more difficult compared to conventional DNA synthesis. The protective group of 2’ hydroxyl in ribose must remain intact both during the synthesis and base deprotection step. The last property of the protection is critical due to the hydrolytic instability of RNA in basic conditions. That is the reason why all modern methods of RNA synthesis that employ different RNA phosphoramidite monomers have protective groups at the 2’ hydroxyl group. This protective group needs to be stable in basic conditions during base deprotection and can be easily removed to completion on the next processing step using different or orthogonal conditions.
III. Synthesis of modified oligonucleotidesThe synthesis of modified oligonucleotides involves the selection of the best strategy to design and plan the needed synthetic approach or pathway. The following paragraph shows how to develop a synthetic strategy and introduce desired modification into oligonucleotide using manual phosphoramidite coupling and how to optimize post synthetic labeling with an activated ester. a) Strategy of synthesis A good way to plan a synthesis is to start with a retro-synthetic analysis. This approach starts with determining what the final product should be and going backwards along the synthetic route to determine what kind of reagents have the appropriate protecting groups needed for the synthesis. An example is shown below. The synthetic path needs to be designed using modifications that do not interfere with other functional groups. That is, the best orthogonal conditions will need to be established. b) On-Support modifications The easiest on support modifications known is the oxidation of H-phosphonates or phosphite triesters using elemental sulfur or disulfides to afford phosporothioates. There are a number of modifications of the phosphate backbone, which can be done while the oligonucleotide is attached to solid support. Most of them require special phosphoramidites, such as the use of alkyl phosphonates and phosphate triesters, or others. Some of them require an H-phosphonate backbone for the transformation. Examples are the use of boranophosphates (Sergueev & Shaw, 1998) or phosphoramidates (Froehler, 1986). Internal modifications on oligonucleotides can be done using either corresponding extendible amidites or using an asymmetrical branch with a non-extendible amidites and any 5’ modifications can be introduced using any type of monomers using an automated synthesizer. In addition any 3’ modification can be introduced using the appropriate solid support. The reversed synthesis from the 5’ to 3’ end can be employed for 3’ modifications if a solid support is not available but the needed phosphoramidite is. The same approach is applicable for 5’ modifications when the modifications are only available on a solid support. Furthermore, phosphoramidite chemistry is not the only chemistry that can be performed using solid support. Some peptide chemistry methods are also compatible with oligonucleotide synthesis methods. For example NHS esters or similar activated esters can be attached to unblocked aliphatic amino-linkers on solid support. In this case the desired molecular probe or reporter molecule has to be stable in basic conditions used during cleavage and base deprotection. c) Post-synthetic modifications. The most common chemo-selective reagents for post synthetic oligonucleotide modifications, for example to introduce different molecular probes and markers, are N-hydroxysuccininidyl activated esters. This type of activated esters reacts very selectively with aliphatic primary amines and is very stable in aqueous buffers at pH 8-9 towards hydrolysis. However, the following three very important issues in post-synthetic NHS labeling need to be addressed. First, the pH of the reaction mixture needs to be maintained at pH 8-9 during the coupling reaction. Second, the correct salt form of the oligonucleotide needs to be used, since ammonia or amino contaminants can also react with NHS esters to form unwanted different by-products. Therefore it is very important to exchange all ammonia ions with other types of counter ions such as lithium or sodium ions. This can be achieved either by precipitation of the oligonucleotides with lithium or sodium ions or by using HPLC with the corresponding buffers. Third, another important requirement for a successful reaction is to make sure that the NHS ester is completely soluble in the reaction mixture of the corresponding buffer and DMSO or other organic solvent suitable for the reaction used. This is needed because the precipitated ester is unreactive. This method can be used not only for conjugation of DNA to small molecules, but also for DNA-protein conjugations (Jablonski et al. 1986). Michael addition is another useful selective reaction for oligonucleotide post synthetic modifications. Maleimide derivatives and sulfhydryl modified oligonucleotides are usually employed for this method. This approach has been used for conjugation DNA and proteins as well (Ransom Hill Bioscience, Inc, Technical Bulletin). There are a few other chemoselective methods, which have been used for DNA–protein conjugations and the attachment of modified oligonucleotides to solid supports, such as those involving aldehydes and hydrazides, disulfides and thioesters, etc. (see IDT Technical Bulletins). All these methods are very useful for the introduction of post-synthetic oligonucleotide modifications. d) Automated DNA/RNA synthesizer One of the earliest, or the earliest, DNA/RNA synthesizer can be viewed at the Smithsonian Institution in Washington DC. The DNA/RNA synthesizer model 394 is made out of steel, glass, plastic and insulated wire. It has the measurements: 50 cm x 66 cm x 46 cm; 19 11/16 in x 26 in x 18 1/8 in.
Figure 22 shows pictures of typical reaction columns containing either activated or monomer loaded CPG supports as well as pictures of automated oligonucleotide synthesizers. Using this type of supports succinylated oligonucleotides or their monomers can be coupled to aminophenyl- or aminopropyl-derivatized glass surfaces, and disulfide-modified oligonuclotides or their monomers can be immobilized onto a mercaptosilanized glass support using a thiol/disulfide exchange reaction or through chemical cross-linkers. Figure 22: Icons of reaction columns and automated oligonucleotide synthesizers are shown. (Left) Different types of columns, color coded. (Middle) Columns mounted into the holder of the synthesizer. (Right) ABI 3900 and Expedite High-Throughput DNA/Oligo Synthesizers. Table 1: Some milestone in DNA/RNA solid phase oligonucleotide synthesis.
Reference Abbreviations and Symbols for the Description of Conformations of Polynucleotide Chains. IUPAC-IUB Joint Commission on Biochemical Nomenclature (JCBN): Recommendations 1982. www.chem.qmul.ac.uk/iupac/misc/pnuc1.html Tatyana Abramova; Review: Frontiers and Approaches to Chemical Synthesis of Oligodeoxyribonucleotides. Molecules 2013, 18, 1063-1075; doi:10.3390/molecules18011063. Avery, OT, MacLeod, CM and McCarty, M (1944). Studies on the chemical nature of the substance inducing transformation of Pneumococcal types. J. Exp. Med. 79, 137-158. Willi Bannwarth; Synthesis of Oligodeoxynucleotides by the Phosphite-Triester Method Using Dimer Units and Different Phosphorous-Protecting Groups. Helvetica Chimica Acta, Volume 68, Issue 7, pages 1907–1913, 13 November 1985. J.C. Biro, B. Benyó, C. Sansom, Á. Szlávecz, G. Fördös, T. Micsik, and Z. Benyó; A common periodic table of codons and amino acids. Biochemical and Biophysical Research Communications 306 (2003) 408–415. Marvin H. Caruthers; The Chemical Synthesis of DNA/RNA: Our Gift to Science. January 11, 2013 The Journal of Biological Chemistry, 288, 1420-1427. F Eckstein. Oligonucleotides and analogues: A practical approach. Edited by F Eckstein. Series editors: D. Rickwood and B.D. Hames. IRL PRESS. Oxford University Press. 1991. Froehler, B.C. Deoxynucleoside H-Phosphonate Diester Intermediates in the Synthesis of Internucleotide Phosphate Analogues Tetrahedron Lett. 1986, 27, 5575-5578. P. T. Gilham and H. G. Khorana; Studies on Polynucleotides. I. A New and General Method for the Chemical Synthesis of the C5″-C3″ Internucleotidic Linkage. Syntheses of Deoxyribo-dinucleotides. J. Am. Chem. Soc., 1958, 80, 6212–6222. Hall, R.H., Todd, A., and Webb, R.F. 1957. Nucleotides. Part XLI. Mixed anhydrides as intermediates in the synthesis of dinucleoside phosphates. J. Chem. Soc. 3291-3296. Jablonski, E.; Moomaw, E.W.; Tullis, R.H. & Ruth, J.L. Preparation of oligodeoxynucleotide-alkaline phosphatase conjugates and their use as hybridization probes. Nucl. Acid Res. 1986, 14, 6115-6128. Kabachnik, M.M.; Potapov, V.K.; Shabarova, Z.A. & Prokofiev, M.A. Dokl. Acad. Nauk SSSR 1971, 201, 858-861. H. G. Khorana, G. M. Tener, J. G.Moffatt and E. H. Pol, Chem. Ind., 1956, 1523. H. G. Khorana, W. E. Razzell, P. T. Gilham, G. M. Tener and E. H. Pol; SYNTHESES OF DIDEOXYRIBONUCLEOTIDES. J. Am. Chem. Soc., 1957, 79, 1002–1003. Y. Lapidot, H. G. Khorana; Studies on Polynucleotides. XXVIII. The Specific Synthesis of C3″-C5″-Linked Ribooligonucleotides (4). The Stepwise Synthesis of Uridylyl-(3″ → 5″)-adenylyl-(3″ → 5″)-uridylyl-(3″ → 5″)-uridine. J. Am. Chem. Soc., 1963, 85 (23), pp 3852–3857. Y. Lapidot, H. G. Khorana; Studies on Polynucleotides. XXIX. The Specific Synthesis of C3″-C5″-Linked Ribooligonucleotides(5). Homologous Adenine Oligonucleotides. J. Am. Chem. Soc., 1963, 85 (23), pp 3857–3862. Letsinger, R.L. & Mahadevan, V. J.; Oligonucleotide Synthesis on a Polymer Support. Am. Chem. Soc. 1965, 87, 3526-3527. Letsinger, R.L. & Mahadevan, V. J. ; Stepwise Synthesis of Oligodeoxyribonucleotides on an Insoluble Polymer Support. Am. Chem. Soc. 1966, 88, 5319-5324. McBride, L.J. & Caruthers, M.H. An investigation of several deoxynucleoside phosphoramidites useful for synthesizing deoxyoligonucleotides. Tetrahedron Lett. 1983, 24, 245-248. A. M. Michelson and Alexander R. Todd; Nucleotides part XXXII. Synthesis of a dithymidine dinucleotide containing a 3′: 5′-internucleotidic linkage. J. Chem. Soc., 1955, 2632-2638. DOI: 10.1039/JR9550002632. J. G. Moffatt, H. G. Khorana; D-XYLOSE-3-PHOSPHATE. J. Am. Chem. Soc., 1956, 78 (4), pp 883–884. Operation Manual. MilliGen/Biosearch Cyclone™ Plus DNA Synthesizer. Usman, N.; Ogilvie, K. K.; Jiang, M. Y.; Cedergren, R. J.; The automated chemical synthesis of long oligoribuncleotides using 2'-O-silylated ribonucleoside 3'-O-phosphoramidites on a controlled-pore glass support: synthesis of a 43-nucleotide sequence similar to the 3'-half molecule of an Escherichia coli formylmethionine tRNA. J. Amer. Chem. Soc. 1987, 109 (25): 7845–7854 D. H. Rammler , H. G. Khorana; Studies on Polynucleotides. XX.1 Amino Acid Acceptor Ribonucleic Acids (1). The Synthesis and Properties of 2″ (or 3″)-O-(DL-Phenylalanyl)-adenosine, 2″ (or 3″)-O-(DL-Phenylalanyl)-uridine and Related Compounds. J. Am. Chem. Soc., 1963, 85 (13), pp 1997–2002. Reese, C.B. & Tompson, E.A.; A new synthesis of 1-arylpiperidin-4-ols. J. Chem. Soc. Perkin Trans. I, 1988, 2881-2885. Roy, S. and Caruthers, M.; Synthesis of DNA/RNA and their analogs via phosphoramidite and H-phosphonate chemistries. Molecules 2013, 18, 14268-14284. The RNA World. Second Edition. Ed. R. F. Gesteland, T. R. Cech, J. F. Atkins. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, New York. Monograph 37, 1999. Subhadeep Roy and Marvin Caruthers; Synthesis of DNA/RNA and Their Analogs via Phosphoramidite and H-Phosphonate Chemistries. Molecules 2013, 18, 14268-14284; doi:10.3390/molecules181114268. Scaringe, S.A.; Wincott, F.E. & Caruthers, M.H.; Preparation of 5′‐Silyl‐2′‐Orthoester Ribonucleosides for Use in Oligoribonucleotide Synthesis. J. Am. Chem. Soc. 1998, 120, 11820-11821. H. Schaller, G. Weimann , B. Lerch , H. G. Khorana; Studies on Polynucleotides. XXIV. The Stepwise Synthesis of Specific Deoxyribopolynucleotides (4). Protected Derivatives of Deoxyribonucleosides and New Syntheses of Deoxyribonucleoside-3″ Phosphates. J. Am. Chem. Soc., 1963, 85 (23), pp 3821–3827. H. Schaller, H. G. Khorana; Studies on Polynucleotides. XXV. The Stepwise Synthesis of Specific Deoxyribopolynucleotides (5). Further Studies on the Synthesis of Internucleotide Bond by the Carbodiimide Method. The Synthesis of Suitably Protected Dinucleotides as Intermediates in the Synthesis of Higher Oligonucleotides. J. Am. Chem. Soc., 1963, 85 (23), pp 3828–3835. H. Schaller, H. G. Khorana ; Studies on Polynucleotides. XXVII. The Stepwise Synthesis of Specific Deoxyribopolynucleotides (7). The Synthesis of Polynucleotides Containing Deoxycytidine and Deoxyguanosine in Specific Sequences and of Homologous Deoxycytidine Polynucleotides Terminating in Thymidine. J. Am. Chem. Soc., 1963, 85 (23), pp 3841–3851. Sergueev, D.S. & Shaw, B.R.; H-Phosphonate Approach for Solid-Phase Synthesis of Oligodeoxyribo-nucleoside Boranophosphates and Their Characterization. J. Am. Chem. Soc. 1998, 120, 9417-9427. Smith, M., Rammler, D.H., Goldberg, I.H. and Khorana, H.G. (1962). Studies on Polynucleotides. XIV. Specific Synthesis of the C3'-C5' Interribonucleotide Linkage. Synthesis of Uridylyl-(3'->5')-Uridine and Uridylyl-(3'->5')-Adenosine. J. Amer. Chem. Soc. 84, 430-440. Smith, M. and Khorana, H.G. (1958). Nucleoside polyphosphates. VI. An improved and general method for the synthesis of ribo-and deoxyribo-nucleoside-5' triphosphates. J. Amer. Chem. Soc. 80, 1141-1145. Sproat BS.; RNA synthesis using 2'-O-(tert-butyldimethylsilyl) protection. Methods Mol Biol. 2005;288:17-32. G. M. Tener, H. G. Khorana, R. Markham, E. H. Pol; Studies on Polynucleotides. II.1 The Synthesis and Characterization of Linear and Cyclic Thymidine Oligonucleotides. J. Am. Chem. Soc., 1958, 80 (23), pp 6212–6222. G. Weimann, H. Schaller , H. G. Khorana; Studies on Polynucleotides. XXVI. The Stepwise Synthesis of Specific Deoxyribopolynucleotides (6). The Synthesis of Thymidylyl-(3″ → 5″)-deoxyadenylyl-(3″ → 5″)-thymidylyl-(3″ → 5″)-thymidylyl-(3″ → 5″)-thymidine and of Polynucleotides Containing Thymidine and Deoxyadenosine in Alternating Sequence. J. Am. Chem. Soc., 1963, 85 (23), pp 3835–3841. Winnacker, Ernst L.; From genes to clones: introduction to gene technology. VCH, 1987. R. S. Wright, H. G. Khorana; Phosphorylated Sugars. I. A Synthesis of β-D-Ribofuranose 1-Phosphate. J. Am. Chem. Soc., 1956, 78 (4), pp 811–816. |
In recent times both basic and applied molecular biology studies have made extensive use of modified oligonucleotides as tools in the study of gene regulation, drug discovery and diagnostics. In particular, aldehyde modified oligonucleotides have been used in applications such as bio-conjugation1 and micro-array2,3 in hybridization-based assays.
The conjugation reaction between aldehyde and hydrazides, hydrazines, semicarbazides or aminooxy fynctionalized synthons is highly chemically selective and efficient.1-3 It was shown that the resulting Schiff-base is relatively stable linker,2 however it can be stabilized further by reduction to the corresponding hydrazine.4 Most methods are orthogonal to the most popular conjugation methods utilizing Michael addition reaction or activated carboxylates. Therefore the aldehyde functional group will not interfere with those reactions and can be used, when other functional groups involved in the parallel process.
Bio-synthesis offers not only wide varieties of 3’-, 5’- and internally aldehyde modified oligonucleotides and also their conjugates with peptides, proteins and antibodies.1
References:
1. http://www.biosyn.com/Bioconjugation.aspx
2. M. A. Podyminogin, E. A. Lukhtanov and M. W. Reed Nucleic Acid Res., 2001, 29, 5090-5098.
3. S. Raddatz, J. Mueller-Ibeler, J. Kluge, L. Wäß, G. Burdinski, J. R. Havens, T. J. Onofrey, D. Wang, and M Schweitzer Nucleic Acid Res., 2002, 30, 4793-4802.
4. E. N. Timofeev, A. D. Mirzabekov, S. V. Kochetkova and V. L. Florentiev Nucleic Acid Res., 1996, 24, 3142-3148.
Epigenetics and the inheritance of epigenetic defectsHave you ever wondered if where you grew up and where you lived or live now, what you eat and what you do had or has an influence on your health? Evidence is mounting that this is the case. Our genes seem to have a memory of our past. Research indicates that the nutritional fate of our grant parents may have left their mark on our genes. How can this be? Bee keepers know how to tell a worker bee from a queen bee. But what makes a worker bee look different from a queen bee? It’s her sister. Unlike in humans, royalty is not inherited in honey bees. The honeybee queen bee and worker bees are genetically identical but the queen larvae get feed a special diet – royal jelly – in large quantities and for long periods to make the difference. The worker bees become slaves to the queen. Something is happening on top of the bee’s genome. Alas, the epigenome came to pass. The honeybee’s epigenome has been studied by Lyko et al. recently and the methylome, the methylated genome, of the brains of worker bees and queen bees has been published in 2010. The pictures below show the two different bees and a depiction of the methylome.
Figure 1: A picture of a worker bee (left), the queen bee (middle) and an icon of the methylome is depicted here. In a paper published in the journal Science in the year 1987 Robin Holliday reports that there is plenty of evidence now originating from many different sources that indicating that control of gene expression in higher organisms is related to the methylation of cytosine in DNA. Furthermore, the pattern of methylation appears to be is inherited. Scientists observed that the loss of methylation, for example resulting from DNA damage, will lead to heritable abnormalities in gene expression. Robin Holliday noted that these events may be important in oncogenesis and aging. Robin further proposed that epigenetic defects in germ line cells due to loss of methylation can be repaired by recombination during meiosis but that some maybe transmitted to offsprings. The term “epigenetics” was coined by Waddington (1942) to refer to the study of the “causal mechanisms” by which “the genes of the genotype bring about phenotypic effects.” Furthermore he stated that “Epigenetics has different meanings for different scientists”. In the mind of a molecular biologist that may involve the study of heritable changes of the DNA that can be observed during mitosis that cannot be explained by changes in DNA sequence. These changes include DNA methylation, histone modification and others. For other scientists epigenetics could refer to interactions of cells and cell products that lead to morphogenesis and differentiation. Simply speaking, epigenetic is the study of genetic effects on the phenotype that are not caused by alteration of the DNA sequence including heritable effects on a genes or chromosomes function that is not accompanied by a change in the DNA sequence. As we will see the notion of epigentics has a long history. However, only in the last 25 years did it become an intensely studied scientific problem waiting to be solved. The figure below shows the explosive increase in publications for epigenetics in general and the epigenetics related to cardiovascular disease from 1995 to 2010. Due to the immense list of publications in the field only some major representative papers will be reviewed here.
Figure 2: Illustration of some epigenetic mechanisms
Figure 3: Increase in research publications covering epigentics since 1995.
What happens when cells develop into different cell types using the same genome as a template? Let us first review what we know so far: Human life begins with a single cell. This embryonic “stem cell” contains all the genetic information needed to develop into a full-grown adult, the genome. Through repeated cell divisions the cell eventually multiplies into ten of trillions of cells. Each cell contains a complete copy of the genome. However, despite having the same genetic information these cells develop into hundreds of different cell types that make up the human body. To find out how this works the field of epigenetics came to pass. A process called mitosis splits a single cell into two cells with identical genetic information. Each cell is capable to develop into different cell types, e.g. into blood cells, neurons or others. We now think that the epigenome determines what type of cell a stem cell will become. Each cell has the same copy of the instruction manual but a brain cell, for example, may only use certain chapters, say to build synapses. The epigenome of each cell tells the cell what chapter to read. Further, we also know now that DNA coils around proteins called histones forming the nucleosome. The helical DNA double strands are packed tightly around the histones inside the nucleus of the cell. The nucleosome is coiled further into a rope like structure called chromatid, which itself is packaged into the chromosome. The epigenome controls access to the genes by attaching molecular caps called methyl groups at certain points of the genes to block them. Histones can coil so tightly around the DNA so that some genes become unreadable. Methyl groups attached to base pairs of a gene changes the expression of the gene. The result is that the DNA of the cells is identical but the epigenetic counterpart is not. Cells perform different functions because now they have different patterns of methyl groups and histones controlling which genes are expressed. DNA methylation is a stable, epigenomic mark occurring at cytosine nucleotides often found within promoter sequences. Histone deacetylases (HDAC) are enzymes that remove acetyl groups (O=C-CH3) from ε-N-acetyl lysines on a histone. The removal of the acetyl group now causes the histones to wrap the DNA around themselves more tightly. DNA methylation is therefore associated with a stable suppression in gene transcription and represents a mechanism to turn genes off. So what exactly is epigenetics?Epigenetics is defined as the study of
Adrian Bird reports in 2007 in “Perceptions in Epigenetics” that there are two classic epigenetic systems. A. There is the Polycomb and Trithorax (Polycomb/Trithorax) system, and B. DNA methylation. The Polycomb and Trithorax groups of proteins, named after mutants of the fruitfly Drosophila melanogaster, maintain repressed or active transcription states, respectively, of developmentally important genes. If they are absent the genes that specify the different segments of the fruitfly are initially expressed correctly but the gene pattern cannot be maintained. It appears that the Polycomb/Trithorax systems establish stable ’memorized’ gene-expression patterns that have been set up by other cellular mechanisms. It has been reported that Polycomb-imposed silencing can even be transmitted between fruitfly generations at low frequencies. Components of the two key Polycomb-system protein complexes have been identified and a close link with modification of the lysine residue at position 27 of histone H3 has been established. However, the mechanism by which silencing is transmitted between cell generations remains unknown. In vertebrates the methylated sequence is CG, which is paired with the same sequence on the opposite DNA strand. Methylation sites are transiently found on only one of the two DNA strands after DNA replication. CG methylation patterns can be copied between cell generations by the DNA methyltransferase DNMT1, which ‘completes’ hemimethylated but not unmethylated sites. DNA methylation is associated with stable gene silencing (for example, on the inactive X chromosome), either through interference with transcription-factor binding or through the recruitment of repressors that specifically bind sites containing methylated CG. Nuclear receptors can transduce environmental and metabolic signals into alterations in gene expression. The recruited coregulator molecules alter the structure of the chromatin. The following figure shows locations of CpG islands in a hypothetical gene. Methylated and unmethylated islands are depicted.  Figure 5: Locations of CpG islands in a hypothetical gene. Methylated and unmethylated islands are shown here. In the year 2000 Allis and Strahl introduced the notion of the “Histone Code”. The researchers reported that a diverse array of post-translational modifications that often occur on tail domains of histone proteins has been well documented. Furthermore, in their paper they proposed that “distinct histone modifications, on one or more tails, act sequentially or in combination to form a 'histone code' that is read by other proteins to bring about distinct downstream events.” The conventional thinking at the time was that the modifications on the histones just strengthened or loosened the nucleosomes hold on DNA altering gene expression accordingly. The histone code hypotheses proposed instead that these epigenetic modifications represent a histone language that other proteins could read, write and erase and modify. If scientist could decipher this code they could predict events such as transcription, chromatin remodeling and silencing. In 2001 Jenuwein and Allis published a paper reporting on how to translate the histone code. Chromatin in all eukaryotes contains an array of posttranslational modifications. The majority of which are found on the amino-termini of histones. They proposed that histone proteins and their associated covalent modifications contribute to a mechanism that can alter the structure of chromatin. These modifications then lead to an inherited difference in transcriptional states. They can be viewed as “off” and “on” states that define the higher order structure of the centromers. Chromatin based events can lead to either gene activation or gene silencing. Differences in histone modifications are descriped as“euchromatic” (“on”) or “heterochromatic” (“off”). A schematic representation of the proposed model is depicted in figure 6.
Figure 6: Proposed model of euchromatin and heterochromatin (2001 Jenuwein and Allis) illustrating the “off” and “on” states that define the higher order structure of the centromers. In the nuclei of all eukaryotic cells, genomic DNA is found as a highly folded, constrained and compacted dynamic polymer. The DNA polymer strands are wound around histones and are compacted further with non-histone proteins. Chromosomal regions that remain transcriptionally inert are highly condensed in the interphase nucleus and can be observed in the cell as heterochromatic foci or as the “Barr body,” which represents the inactive X chromosome in female mammalian cells. Roughly two superhelical turns of DNA wrap around an octamer of core histone proteins in the nucleosome. The core histone proteins that form the octamer are H3-H4, found as a tetramer, and H2A-H2B dimers. Further studies will be needed to find out how the addition of the linker histone 1 (H1) protein causes the chromatin fiber to form a more compacted filament with a defined higher ordered structure. Covalent modifications such as acetylation, methylation, and phosphorylation are found on the histone tails allowing for regulatory contacts with the underlying DNA. Highly specific enzymes that converts these histone tail modifications have been identified and the list of them is getting longer by the day. The histone code hypothesis predicts that
Available experimental data link alterations in chromatin structure to cell cycle progression, DNA replication, DNA damage and repair, recombination and chromosome stability.
Figure 7: Examples of combinatorial modifications in the histone amino acid termini representing imprints for active or inactive chromatin. The field of epigenetics has the potential to revolutionize the field of medical research and healthcare. It encompasses the study of nuclear components such as chromatin structure, including histone modifications, protein/DNA interactions, protein/RNA interactions, and how these factors influence gene function. It also includes the study of DNA methylation and the role that non-coding RNAs (ncRNAs) play in influencing DNA methylation patterns, chromatin structure and the regulation of gene expression. The polymerase chain reaction (PCR) can be used to study DNA methylation patterns, densities, and even the methylation status of individual cytosine residues. In addition, PCR methods have been developed to survey ncRNA expression and to identify regions of the genome where proteins and RNA interact or where certain functional histone marks are located. ReferencesAdams, J. (2008) Obesity, epigenetics, and gene regulation. Nature Education 1(1) Adrian Bird. Introduction Perceptions of epigenetics Nature 447, 396-398 (24 May 2007) Dolinoy, D. C., et al. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proceedings of the National Academy of Sciences 104, 13056–13061 (2007) Choudhuri S. Obesity, Epigenetics, and Gene Regulation By: Jill U. Adams, Ph.D. (Freelance Science Writer) © 2008 Nature Education Choudhuri S. From Waddington's epigenetic landscape to small noncoding RNA: some important milestones in the history of epigenetics research. Toxicol Mech Methods. 2011 May;21(4):252-74 Duhl, D. M., et al. Neomorphic agouti mutations in obese yellow mice. Nature Genetics 8, 59–65 (1994). Andrew P Feinberg. Methylation meets genomics. Nature Genetics 27, 9 - 10 (2001) D. Haig. “The (Dual) Origin of Epigenetics” 2004, Cold Spring Harbor Symposia on Quantitative Biology Volume LXIX. Holliday R. The inheritance of epigenetic defects. Science. 1987 Oct 9;238 (4824):163-70. Holliday, Robin. “Epigenetics: A Historical Overview.” Epigenetics 1(2006): 76-80. Lu, D., et al. Agouti protein is an antagonist of the melanocyte-stimulating-hormone receptor. Nature 371, 799–802 (1994). Adele Murrell, Vardhman K. Rakyan and Stephan Beck; From genome to epigenome. Human Molecular Genetics, 2005, Vol. 14, Review Issue 1. Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000 Jan 6;403(6765):41-5. Venter et al., 291 (5507): 1304-1351. The Sequence of the Human Genome Science 16 February 2001: Vol. 291 no. 5507 pp. 1304-1351. Robert A. Waterland and Randy L. Jirtle. Transposable Elements: Targets for Early Nutritional Effects on Epigenetic Gene Regulation. Mol. Cell. Biol. August 2003 vol. 23 no. 15 5293-5300 Resources for EpigeneticsConsortia and InitiativesThe NIH Roadmap Epigenomics Program: A NIH Initiative to foster epigenomic research, develop comprehensive reference epigenome maps, and generate new technologies for comprehensive epigenomic analyses. http://nihroadmap.nih.gov/epigenomics/ The Epigenome Network of Excellence: An EU-funded network of institutions and research groups http://www.epigenome-noe.net/WWW/index.php The Human Epigenome Projects: A public/private collaboration to catalogue Methylation Variable Positions (MVPs) in the human genome NAME21: A German National Initiative to analyze DNA methylation Patterns of Genes on Chromosome 21 http://biochem.jacobs-university.de/name21/ DatabasesThe Human Epigenome Atlas: The atlas includes human reference epigenomes and the results of their integrative and comparative analyses. http://www.genboree.org/epigenomeatlas/index.rhtml MethDB: A searchable database for DNA methylation and environmental epigenetic effects Human Histone Modification Database (HHMD): A searchable database of information from experimental data to facilitate understanding of histone modifications at a systematic level. The current release incorporates 43 location-specific histone modifications in human. http://bioinfo.hrbmu.edu.cn/hhmd NCBI Epigenomics: An online repository of epigenetic datasets http://www.ncbi.nlm.nih.gov/epigenomics/browse GeneImprint: A catalogue of imprinted genes http://www.geneimprint.com/site/genes-by-species Catalogue of Parent of Origin Effects: Searchable database of imprinted genes and related effects http://igc.otago.ac.nz/home.html Tools and Other ResourcesMethPrimer: Primer Design for Methylation PCR http://www.urogene.org/methprimer/index1.html MethBlast: A sequence similarity program that checks your primers for bisulfite converted DNA by blasting them against unmethylated and methylated genomic sequences of man, mouse and rat http://medgen.ugent.be/methBLAST Methylator: Methylator attempts to predict whether CpGs in a DNA sequence are likely to be methylated or not http://bio.dfci.harvard.edu/Methylator/ RMAP: RMAP is a tool to map reads from the next-generation sequencing technology that supports bisulfite-treated reads mapping. Chromatin Structure & Function: Information on chromatin biology, histones and epigenetics http://www.chromatin.us/chrom.html Epigenetic Station: A source for information, protocols, methods, techniques, products, vendors, kits, assays, analysis, bioinformatics and databases on Epigenetics |
The use of biological therapies for the treatment of cancer is now a fast growing field. Unfortunately, the use of naked antibodies that show clinical efficacy has proven to be limited. To enhance the therapeutic potential of an antibody the conjugation to a small drug molecule has been heavily investigated in recent years. This approach is expected to combine the benefits of highly potent drugs with selective binders of specific tumor antigens. In addition, the next generation of ADCs are designed to utilize site-specific coupling chemistries using protein engineering with the aim to maintain the specificity and potency but to also improve the half-life, the homogeneity of drug loading, safety and quality of the therapeutic molecule. However, designing an ADC is a complex undertaking that requires the thoughtful combination of a selected antibody with a chemical linker group and a drug that targets a defined cancer indication. Lessons learned from the first-generation antibody-drug conjugates resulted in the improvement of the technology that now guides the design of new improved compounds, some of which are now in clinical trials. Intense research is ongoing to help improve ADCs further. Various linkers that show good in vitro stability and have a positive effect on ADC PK and in vivo efficacy are now designed and investigated.
As of 2013 a total of 27 ADC were undergoing clinical trials in both hematological malignancies and solid tumor indications. One ADC called T-DM1 or trastuzumab emtansine showed very promising results in phase III for the treatment of HER2-positive refractory/relapsed metastatic breast cancer. Other compounds currently in clinical trials are CMC-544, SAR3419, CDX-011, PSMA-ADC, BT-062, and IMGN901, targeting varied antigens. In additions, a search performed in April 2014 at “ClinicalTrials.gov” for the term “antibody-drug-conjugates” showed that 213 clinical trials were ongoing and some of them are recruiting.
Since the ADCs are exposed to different conditions during their journey from the blood vessels to the molecular target in the tumor tissue the mode of action of these drugs at the cellular or molecular level is complex. During circulation in the plasma the ADC must behave like a natural antibody and the linker must be stable. During antigen binding it is necessary that the conjugated antibody retains high immunoaffinity and the attached drug compound must not disturb this. During internalization a sufficient intracellular concentration of the drug must be achieved. After internalization during drug release the ADC has to efficiently release the original cytotoxic drug in its active form in the tumor. And finally, the inherent potency of the released drug must be sufficient to kill the tumor cells. These criteria demand that the ADCs are properly designed to achieve optimal effects.
References and Links
By: Klaus D. Linse
Collagen mimetic peptides, or CMPs, are typically made of 30 or fewer amino acids. These types of peptides are usually composed of multiple helix promoting peptide trimers. Collagens are integral structural proteins which are among the most diverse and abundant proteins found in the animal kingdom where they play key functional roles in cellular modulation. Therefore these proteins have attracted scientists in the research fields of supramolecular chemistry, biomedical and materials science in recent years as a guide for the design of unique synthetic biomolecules. In the last three decades CD4 α-turn mimetic peptides that inhibit human immunodeficiency virus envelope glycoprotein gpl20 binding and infection of human lymphocytes have been designed and synthesized. In addition the design of similar peptide mimics has blossomed as well. Collagen mimetic peptides were initially developed and used by biochemists for the investigation and elucidation of the structures and stability of natural collagens. Biologists and polymer chemists followed soon to produce nanostructured fibrous scaffolds using collagen mimetic peptides as the building blocks. The design of CMPs is based on ProProGly and ProHypGly trimer sequence motifs. The best characterized CMPs to date contain the collagen-like triple-helical structure within their peptide sequence and show reversible melting characteristics that are well documented in the literature. Over the years modern synthesis methods have been developed employing techniques such as ligation chemistries based on activated esters, click chemistry, carbodiimide chemistry or other ligation chemistries. These methods now provide synthetic scientists versatile strategies to prepare collagen-polymer conjugates. Furthermore, researchers observed that these collagen mimetic peptide conjugates can spontaneously assemble when stimulated accordingly. Development of engineered tissue and organ replacement therapies has increased in recent years, promoting a emand for new approaches to immobilize components derived from outside cells or tissue to natural collagen.
These peptide mimics have similar behavior as amphiphilic peptides that are known to form defined nanostructures such as molecular wires, well defined nanotubes as well as nanovesicles. Amphiphilic peptides have been used as scaffolds for the synthesis of defined nanometer structures in recent years and studies of biological systems on the molecular level in the 20th century revealed that molecular self-assembly is a fundamental process in all living systems.
Wang et al. in 2004 developed an alternative to the conventional “covalent” modification methods called a “physical” modification technique that is based on collagen’s native ability to associate into a triple-helical molecular architecture. Chemical coupling of synthetic moieties to amino acid side chains such as lysines (K, Lys) or glutamines (E, Glu) is a routinely used technique for such purposes. Unfortunately, these types of coupling reactions are difficult to control when used on large proteins and generally are not easy to control when modifying integrated collagen scaffolds that contain live cells and tissues. To circumvent this the research group synthesized collagen mimetic peptides containing the sequence -(Pro-Hyp-Gly)- multiple times. The scientists report that these peptides exhibit a strong affinity to both native and gelatinized type I collagen under controlled thermal conditions. Furthermore, they show that the cell adhesion characteristics of collagen can be readily altered by applying a poly (ethylene glycol)-CMP conjugate to a prefabricated collagen film. The next table shows the melting behavior of selected peptides.
Table 1: Melting Transition Temperatures of Collagen Mimetic Peptide Derivatives Determined by Circular Dichroism Spectroscopya (Source: Wang et al., 2005).
Compound | Sequence | Tm (°C) |
1 | -(ProHypGly)10- | 69 |
2 | 5CF-Gly3-(ProHypGly)10- | 75 |
3b
| 5CF-Gly3-randomPro10Hyp10Gly10 |
|
4 | 5CF-Gly3-(ProHypGly)6- | 25 |
a Measured in 57.5 µM acetic acid solution.
b 5CF-GGGGPPPHPHGPGGG PPHPPHGPHGPPHPGPHPHPGGPHPHPP, (PH:Hyp).
c mPEG2000, CH3O-(CH2-CH2-O)n-OH, 2250 Da.
The researchers demonstrated the binding of the synthetic CMP to natural acid soluble, bovine type I collagen or denatured gelatin collagen by treating collagen films with solutions of a fluorescently labeled CMP. Results from rinsing the treated collagen films and measuring the fluorescence intensity of the exposed film suggested that 5-carboxy fluorescein (5CF)-labeled-Gly3-(ProHypGly)
In 2007 Rele et al. designed and synthesized collagen-mimetic triple helix promoting peptides that self-assembled into a fibrous structure with well-defined periodicity as visualized by transmission electron microscopy (TEM). The researchers used a Xaa-Yaa-Gly triad sequence to create sequence specific peptides containing three different Xaa-Yaa-Gly domains, including a central core of Pro-Hyp-Gly repeat sequences flanked by distinct sets of peptide repeats, containing either negatively (Glu) or positively (Arg) charged amino acid residues. The Pro-Hyp-Gly peptide sequence was reported to form the structurally critical hydrophobic core of the assembly, responsible for maintaining the thermodynamic stability of the collagen triple-helical structure. Furthermore, the researchers reasoned that the synthesis of collagen-mimetic triple helix peptide protomers (THPs) that display the capacity to form triple helices with improved stability and that exhibit a propensity to form linear assemblies through a process of axially oriented alignment will prove to have a number of important practical applications in the design of novel biomaterials. These types of material may lend themselves for the development of collagen-based biomaterials for wound healing.
In 2008 Cejas et al. synthesized collagen model peptides that form triple helices and self-assemble into supra-molecular fibrils exhibiting collagen-like biological activity without the need for preorganizing the peptide chains by covalent linkages. The researchers accomplished this by placing aromatic groups on the ends of a representative 30-mer CMP, (GPO)10, by using L-phenylalanine and L-pentafluorophenylalanine in 32-mer. The use of atomic force microscopy topographical imaging indicated that some of these peptides self-organized into microfibrillar species. In addition, two peptides, 1a and 1b, where reported to induce the aggregation of human blood platelets with a potency similar to type I collagen.
Su et al. in 2010 demonstrated that treatment with the apoA-I mimetic peptides, L-4F, D-4F (the peptide Ac-D-W-F-K-A-F-Y-D-K-VA-E-K-F-
Yu et al. in 20011 reviewed progress made in the field of collagen mimetic peptides that are useful for the design and synthesis engineered collagen-like materials for potential biomedical use. The scientists report that the collagen triple helix has become a promising structural motif for engineering self-assembled, hierarchical constructs similar to natural tissue scaffolds. Further, they discuss various CMPs and collagen-like proteins that mimic either structural or functional characteristics of natural collagens. This paper provides helpful information to bioengineers and biomaterials scientists interested in collagen engineering.
Li et al. in 2012 reported the synthesis and use of collagen mimetic peptides (CMPs) that can be phototriggered to fold into triple helix and bind to collagens denatured by heat or by matrix metalloproteinase (MMP) digestion. The peptide binding assays that were used by this research group indicated that the binding is primarily driven by stereo-selective triple-helical hybridization between monomeric CMPs of high triple-helical propensity and denatured collagen strands. Furthermore the scientists showed that photo-triggered hybridization allows specific staining of collagen chains in protein gels as well as photo-patterning of collagen and gelatin substrates. Their in vivo experiments demonstrated that systemically delivered CMPs can bind to collagens in bones, and in articular cartilages and tumors characterized by high MMP activity. They further showed that CMP-based probes can detect abnormal bone growth activity in a mouse model of Marfan syndrome. This approach allowed the researcher targeting the microenvironment of abnormal tissues.
He et al. in 2013 reviewed modern synthesis methods that were developed for the synthesis of collagen mimetic peptide conjugates used in polymer science. These methods employ particular ligation chemistries basing on activated ester, click chemistry, carbodiimide chemistry or other ligation chemistries allowing the preparation of collagen-polymer conjugates. Furthermore, the researchers point out that these conjugates made with collagen mimetic peptides as the building blocks show exciting stimuli responsive or spontaneously assembly behavior.
All these findings have let researchers in the tissue engineering and biomedical field now to speculate that the ability to control the organization of cells in collagen matrices may provide new pathways to engineer new types of tissues. Furthermore, the affinity between the CMPs and collagen could be used to immobilize therapeutic drugs to collagens in living tissues and biomaterials that incorporate natural collagens.
To conclude, highly helical peptides can now be made using a variety of peptide sequences. These types of peptide mimics may have many applications in experimental biology, biomedicine and tissue engineering. Since many peptides can retain ligand-binding properties of proteins from which they are derived they may act as inhibitors of antigen-antibody reactions or of hormone-receptor interactions, which would make them good starting molecules for the design of new types of biomaterials.
References
Mabel A. Cejas, William A. Kinney, Cailin Chen, Jeremy G. Vinter, Harold R. Almond, Jr., Karin M. Balss, Cynthia A. Maryanoff, Ute Schmidt, Michael Breslav, Andrew Mahan, Eilyn Lacy, and Bruce E. Maryanoff; Thrombogenic collagen-mimetic peptides: Self-assembly of triple helix-based fibrils driven by hydrophobic interactions. PNAS June 24, 2008 vol. 105 no. 25, 8513–8518.
SHAOXING CHEN, R. ALAN CHRUSCIEL, HIROSHI NAKANISHIt, APICHAYA RAKTABUTR,
MICHAEL E. JOHNSON, ALICE SATO, DAVID WEINER, JIM HOXIE, HORACIO URI SARAGOVI, MARK I. GREENE, AND MICHAEL KAHN; Design and synthesis of a CD4 a-turn mimetic that inhibits human immunodeficiency virus envelope glycoprotein gpl20 binding and infection of human lymphocytes. Proc. Natd. Acad. Sci. USA Vol. 89, pp. 5872-5876, July 1992.
Lirong He, Patrick Theato, Collagen and collagen mimetic peptide conjugates in polymer science. European Polymer Journal, Volume 49, Issue 10, October 2013, Pages 2986–2997.
Yang Li, Catherine A. Foss, Daniel D. Summerfield, Jefferson J. Doyle, Collin M. Torok, Harry C. Dietz, Martin G. Pomper, and S. Michael Yu; Targeting collagen strands by photo-triggered triple-helix hybridization. 10.1073/pnas.1209721109 PNAS September 11, 2012 vol. 109 no. 37 14767-14772.
Shyam Rele, Yuhua Song, Robert P. Apkarian, Zheng Qu, Vincent P. Conticello, and Elliot L. Chaikof; D-Periodic Collagen-Mimetic Microfibers. J. AM. CHEM. SOC. 2007, 129, 14780-14787.
Feng Su, Kathy R. Kozaka, Satoshi Imaizumib, Feng Gaoa, Malaika W. Amneusa, Victor Grijalvab, Carey Nga, Alan Wagnerb, Greg Houghb, Gina Farias-Eisnerb, G. M. Anantharamaiahc, Brian J. Van Lentenb, Mohamad Navabb, Alan M. Fogelmanb, Srinivasa T. Reddya, and Robin Farias-Eisnera; Apolipoprotein A-I (apoA-I) and apoA-I mimetic peptides inhibit tumor development in a mouse model of ovarian cancer. PNAS | November 16, 2010 | vol. 107 | no. 46 | 19997–20002.
Wang AY, Mo X, Chen CS, Yu SM; Facile modification of collagen directed by collagen mimetic peptides. J Am Chem Soc. 2005 Mar 30;127(12):4130-1.
S. Michael Yu, Yang Li and Daniel Kim; Collagen mimetic peptides: progress towards functional applications. Soft Matter, 2011,7, 7927-7938.
Gold nanoparticles (colloidial gold) have been extensively used for applications both in biology (e.g. bio-imaging) and technology (e.g. photonics) due their unique optical properties. These properties are conferred by the interaction of light with electrons on the gold nanoparticle surface. At a specific wavelength (frequency) of light, collective oscillation of electrons on the gold nanoparticle surface cause a phenomenon called surface plasmon resonance resulting in strong extinction of light. The particular wavelength, or frequency, of light where this occurs is strongly dependant on the gold nanoparticle size, shape, surface and agglomeration state as described in more detail below.
The influence of gold nanoparticle size on the surface plasmon resonance is illustrated in figure 1 below where the absorption maximum (lambda max) increases from 520nm to 570nm for 20nm and 100nm spherical gold nanoparticles, respectively. Particles with sizes above 100nm have broader peaks spanning into the 600nm range due to the presence of both transversal and longitudinal surface plasmon resonances. In comparison, gold nanoparticles with diameters below 2nm do not exhibit surface plasmon resonance.
The difference in extinction between different sized gold nanoparticles can conveniently be utilized for multiplexing.
Figure 1. Gold nanoparticle size dependant surface plasmon resonance. Note the red-shift of the absorption maximum as the gold nanoparticle size increases.
A major determinant of the optical properties of gold nanoparticles is their shape. By synthesizing gold nanoparticles of different shapes, the surface plasmon resonance can easily be tuned to give absorption maxima from around 500nm into the near-infrared part of the spectrum. As an example, spherical collodial gold have absorbance maxima between 515-570nm as described above, while irregular shaped particles such as gold nanorods, and urchin shaped gold nanoparticles (also called gold nanostars) have absorption maximum in the near-infrared region of the spectra, figure 2. For custom synthesis of gold nanoparticles with irregular shapes please contact us.
The difference in absorption properties between spherical and irregular-shaped gold nanoparticles of the same average size is caused by an anisotropic (uneven) distribution of the surface electron layers.
Figure 2. (Top) Gold nanoparticle shape dependant localized surface plasmon resonance as indicated by the visual appearance and UV-VIS spectra of spherical (A), and urchin-shaped (B) gold nanoparticles (gold nanostars). (Bottom) Absorbance spectra for gold nanorods with three different aspect ratios. Note the presence of two absorption peaks, which are caused by both transversal and longitudinal surface plasmon resonances.
Urchin shaped (spiky) gold nanoparticles (gold nanostars) are preferable over spherical particles in in vivo based applications due to a reduced background, and higher penetration of near-infrared light through biological tissues. Also, irregular-shaped gold nanoparticles give higher signal in Surface-Enhanced Raman Spectroscopy (SERS) due to enhancement of the electromagnetic field on the surface caused by the irregular shaped particles. In comparison, spherical particles are ideal for use in application such as immunogold dot-blot protocols (see figure 3 below) and lateral flow rapid tests.
Figure 3. Immuno-dot blot assay illustrating the difference in appearance (color) for three different types of noble metal protein conjugates varying in shape and composition.
As mentioned above, the aggregation state of gold nanoparticles have an effect on their optical properties. This fact can be used to monitor gold nanoparticle stability, both over time, and upon addition of salt-containing buffers, which at high enough concentrations cause particle aggregation, figure 4. The red-shift in absorption maximum caused by aggregation, or particles in close proximity, have successfully been utilized in many assays as a detection mechanism.
Figure 4. Visual appearance and UV-VIS spectra of monodisperse (A) and sodium chloride (NaCl) induced agglomeration (B) of 15nm gold nanoparticles.
Store product away from direct sunlight at 4-25°C. Lower temperature prolongs the shelf life of the product. Do NOT freeze. If frozen, the gold nanoparticles will irreversibly aggregate. This is indicated by a change in color of the solution, see figure below. When stored as specified the colloidal gold is stable for at least 1 year.
When stored for a long period of time the gold nanoparticles might sediment at the bottom of the flask, which is especially true for larger particle sizes. Prior to use, re-suspend the sedimented particles by swirling until a homogenous solution is obtained.
To maintain optimal performance, and stability of the colloidal gold, care should be taken to use clean storage containers if using other than supplied with the product.
Although it is not always necessary to wash the gold nanoparticles prior to use, some applications might require additional washing procedures. The easiest way to remove possible contaminants in the nanoparticles solution is by centrifugation. Centrifugation force is dependant on size of the gold nanoparticles and should be adjusted according to table I for optimal performance.
Note:
Since non-functionalized gold nanoparticles are sensitive to salt containing buffers, re-suspension should always be performed in ultra-pure water to prevent irreversible aggregation. Irreversible aggregation is characterized by a clear to bluish solution upon the addition of salt.
Table I. Appropriate G forces for centrifugation of gold nanoparticles. Note that recommended conditions are for a volume of 1ml and centrifugation using a microcentrifuge, except for 5nm gold nanoparticles that requires an ultracentrifuge.
| Size (nm) | Speed (g) | Time (min) |
|---|---|---|
| 5 | 100,000 | 30 |
| 10 | 17,000 | 60 (~50% recovery) |
| 15 | 17,000 | 30 |
| 20 | 6,500 | 30 |
| 30 | 4,500 | 30 |
| 40 | 2,500 | 30 |
| 50 | 2,000 | 30 |
| 60 | 1,125 | 30 |
| 80 | 600 | 30 |
| 100 | 400 | 30 |
| 150 | 180 | 30 |
| 200 | 100 | 30 |
Note: The amount of protein needed to saturate the gold colloid can also be determined by agarose gel electrophoresis due to the change in charge upon binding of the protein.
Thobhani, S., Atree, S., Boyd, R., Kumarswami, N., Noble, J., Szymanski, M., Porter, R.A. (2010)
Bioconjugation and characterization of gold colloid-labelled proteins Journal of Immunological Methods 356, 60-69
When membranes of blotted proteins are probed with a secondary gold conjugate, the presence of a target protein is indicated with a red color upon binding of the gold conjugate.
When combined with silver enhancement (i.e. deposition of silver onto bound gold colloid turning the spot dark in color) sensitivity in western blot and dot-blot applications rivals that of colorimetric detection methods. In addition, secondary gold probes adapt well to standard western blot protocols and little changes are necessary to your current detection scheme.
Figure 1. Example dot-blot assay for streptavidin gold conjugate (top left) and streptavidin silver conjugate (top right) before and after enhancement using silver enhancement kit for membranes. Bottom picture illustrates and highlights the difference in appearance (color) of 50nm anti-human IgG noble metal nanoparticle conjugates prepared using NHS-activated gold nanoparticles, NHS-activated gold nanourchins and NHS-activated silver nanoparticles, respectively.
1. M. Moeremans, et al., Journal of Immunological Methods, 1984, 74, 353
One that can, and is in widespread use as a result, is the lateral flow immunoassay test, also known as the immunochromatography assay, or strip test. Like many of the best ideas, lateral flow immunoassays take clever and sophisticated technology and turn it into something so simple to operate that alsmost anyone can use it.
The basic technology that underlies lateral flow immunoassays was first described in the 1960s, but the first real commercial application was Unipath’s Clearview home pregnancy test launched in 1988. Since then, this technology has been employed to develop a wide and ever-growing range of assays for clinical, veterinary, agricultural, food industry, bio-defence and environmental applications.
Strip assays are extremely versatile and are available for an enormous range of analytes from blood proteins to mycotoxins and from viral pathogens to bacterial toxins. Assays has even been developed for wine producers to assess the amount of botrytis rot in newly harvested grapes as well as for use in the clinical lab identifying cardiac chemistries. This shows the vast range that this technology can be applied too.
Lateral flow immunoassays are essentially immunoassays adapted to operate along a single axis to suit the test strip format. There are a number of variations of the technology that have been developed into commercial products, but they all operate using the same basic principle.
A typical test strip consist of the following components:
The components of the strip are usually fixed to an inert backing material and may be presented in a simple dipstick format or within a plastic casing with a sample port and reaction window showing the capture and control zones.
We provide a full product line of gold nanoparticles (colloidal gold) for use in a variety of lateral flow assays. A diverse product line of different type of nanoparticles offers you products with a narrow size distribution (CV of less than 12%), exceptional adsorption and conjugation properties and with greater than 95% spherical particles. In addition, batch to batch variability is extremely low (+/- 2nm), which assures that customer will always end up with a product within the specified size range that you ordered.
The high shape uniformity of colloidal gold will minimize the variability within your assay by e.g. allowing control over the available surface area while absorbing or covalently conjugating proteins to gold nanoparticles. It will also ensure a more uniform flow rate across your membrane for improved reproducibility and overall results.
PEG is a polyether molecule and is typically described by the molecular weight and whether they are linear, branched, star, or combed-shaped. PEG molecules can also be functionalized with thiols, amines, carboxylic acids, or alcohols. PEG molecules are coated onto gold nanoparticles by a sulfur-gold atom bond. Interestingly, coating of a dense layer of PEG onto gold nanoparticles has shown to reduce non-specific binding of proteins (2). It was recently shown that gold nanoparticles with greater than 0.96 PEG/nm2 is required to reduce non-specific binding and to inhibit their uptake into macrophage cells. Using these results, PEGylated gold nanoparticles were designed with the lowest non-specific cellular uptake. This is very important when using gold nanoparticles in biology where the particles are programmed to target specific molecules or cellular receptors. Non-specific protein binding can affect the specificity. In addition, the gold nanoparticle-PEG system contains protruding carboxylic acids or amines that allows other molecules to be conjugated to the surface by using the coupling agent 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide, better known as EDC. In this reaction, the gold nanoparticles is incubated with EDC and the biological molecule of interest for 2 hrs and then purified by centrifugation to remove excess biological molecules. These gold nanoparticle conjugates are then ready to be used for biological purposes.
Gold nanoparticles cannot recognize specific biological targets without surface modification. It is well known that single-stranded DNA’s called oligonucleotides can recognize a complementary sequence, antibodies recognize antigens, and peptides recognize antibodies. By coating the surface of gold nanoparticles with oligonucleotides, antibodies, peptides, or other bio-recognition molecules, one is then able to recognize specific targets in solution, in western blots (see figure 1), on/in cells, or in tissues in animals. Basically, these molecules provide gold nanoparticle with a biological function. Typically these bio-recognition molecules are coated onto the surface via direct adsorption or by covalent conjugation to the gold nanoparticle surface containing carboxylic acid or amine functional groups.
Figure 1. Immuno-dot blot assay for detection of human IgG antibodies using gold nanoparticles, silver nanoparticles and gold nanourchins ("spiky gold") coated with bio-recognition molecules. Note how the difference in appearance (color) of the dots can be achieved using different types of noble metal protein conjugates varying in either shape or composition.
Engineering a gold nanoparticle surface that contains PEG and bio-recognition molecules is ideal for their application. The PEG molecule prevents non-specific binding while the bio-recognition molecule provides biological specificity. Gold nanoparticle products enables researchers to design their nanoparticles in this manner, for optimum biological use.
Gold nanoparticles coated with PEG or/and bio-recognition molecules (BRM) has many applications, e.g. biosensors, cellular probes, drug delivery vehicles, or as optical contrast agents. Below, are some specific examples of how to use gold nanoparticle conjugates.
Single stranded DNA-coated onto gold nanoparticles can be used for detection of genetic material (3). In this application, single stranded oligonucleotide-coated gold nanoparticles are incubated with a DNA fragment of interest. If the fragment is complementary to the oligonucleotide sequence on the gold nanoparticles, particles are assembled together. As a result of this “aggregation” the color of the solution changes from red to blue because the surface plasmon is coupled when particles are in the aggregated state. The color change thus indicates a positive detection. Mutations can be detected by heating the sample. DNA de-hybridizes when heated and when a mutation is present in the sequence, the melting temperature is lowered. By measuring and comparing the temperature of a mutated sequence to a perfectly complementary sequence, one is able to detect whether a mutation is present or not. More complex schemes can also be designed to identify the location of a mutation within the DNA fragment analyzed.
The gold nanoparticle PEG/BRM system can be designed to selectively target tumors and bind to cancerous cells. Interestingly,PEGylated gold nanoparticles can target tumors by a passive mechanism alone. An advantage of the protective PEG-layer on the gold nanoparticles is that it has been shown to reduce macrophage uptake. Macrophages are part of the reticuloendothelial system that removes foreign materials from the blood. The PEGylated layer reduces the interaction of the gold surface with blood proteins thereby minimizing non-specific macrophage uptake. This allows the nanoparticles to reside in the blood for long-term, which allows for a greater chance of tumor extravasation. If coated with a drug or imaging agent, the gold nanoparticles can be used as a visualization tool as well as a delivery vehicle to the tumor. Another method of targeting tumors is by coating the gold nanoparticles with a bio-recognition molecule that recognizes receptors on tumor cells, the extracellular matrix, or blood vessels, i.e. active targeting. The advantage of using gold nanoparticles is that one can control the delivery efficiency by the size, shape, or surface chemistry (4).
One of the key questions facing researchers is to understand how the size, shape, and surface chemistry (known as the physico-chemical properties) affect how nanoparticles distribute in the cell and body, and whether specific nanomaterial designs cause toxicity. Gold nanoparticles are ideal platforms to perform these studies on because they can be synthesized and tuned with a narrow size distribution. Further, designs with different shapes and surface chemistries can be readily achieved. This allows one to systematically evaluate their behavior in biological systems. By performing such studies, one is then able to identify the designs with the lowest toxicity, which subsequently can be selected for the final application(s).
Gold nanoparticles scatter light, and when using a dark field microscope to image them, they appear as bright spots (similar to fluorescence). A unique advantage is that the dark field signal does not photobleach like fluorescence from dyes. By labeling gold nanoparticles with bio-recognition molecules such as an antibody to a cell surface receptor etc. cells can thus conveniently be targeted and imaged. In addition, low-level target detection can be improved by using silver enhancement of bound gold nanoparticles.
Most applications in biosensing and detection exploit the optical properties of silver nanoparticles, as conferred by the localized surface plasmon resonance effect. That is, a specific wavelength (frequency) of incident light can induce collective oscillation of the surface electrons of silver nanoparticles. The particular wavelength of the localized surface plasmon resonance is dependant on the silver nanoparticle size, shape, and agglomeration state.
Figure 1 shows the plasmon resonance spectra of different sizes of silver nanoparticles, as measured with UV-visible spectroscopy. As the particle size increases from 10 to 100 nm, the absorbance peak (lambda max) increases from 400 nm to 500 nm, and broadens in width. For particles of greater size, especially above 80 nm, a secondary peak at lower wavelength becomes apparent, which is a result of quadrupole resonance, in addition to the primary dipole resonance.
Figure 1. Silver nanoparticle size dependant surface plasmon resonance. Note the red-shift of the absorption maximum as the gold nanoparticle size increases.
The aggregation state of silver nanoparticles also has significant effect on their optical properties. This fact can be used to monitor the stability of silver nanoparticles, both over time, and upon addition of salt-containing buffers, which at high enough concentrations cause particle aggregation. As shown in the absorbance spectra in Figure 2, aggregation is indicated by a decrease in the primary peak, and an increase in the red region.
Figure 2. Visual appearance and UV-VIS spectra of monodisperse (A) and sodium chloride (NaCl) induced agglomeration (B) of 10nm silver nanoparticles.
Both silver nanoparticles and gold nanoparticles are commonly employed in optical detection for their surface plasmon resonance effect. The plasmon excitation efficiency of silver nanoparticles is known to be even more pronounced than that of gold nanoparticles, as shown in their stronger, sharper plasmon resonance peaks at the same particle concentration. Silver nanoparticles thus can render better sensitivity for some applications, such as localized surface plasmon resonance or surface enhanced Raman scattering detection.
Silver nanoparticles may also be advantageous over gold nanoparticles, when used in combination with fluorescence emission detection. Most fluorophores emits at a wavelength above 500 nm. However, the plasmon resonance absorbance of gold nanoparticles is primarily in the range of 500-600 nm, and hence can quench the detectable fluorescence to some extent, when the fluorescent dyes are close to the particle surface. This issue of fluorescence quenching is minimized for silver nanoparticles, as their plasmon resonance absorbance is mostly below 500 nm, with little overlapping with the emission wavelength of most fluorescent dyes.
With their different wavelengths of plasmon resonance, silver and gold nanoparticles, or particles of different size or shapes, can also be used together for multiplexed detection, taking advantage of the extended range of detection spectrum. Figure 4 shows dark field microscope images of silver nanoparticles, gold nanoparticles and gold nano-urchins.
Figure 4. This image highlights the difference in appearance between silver nanoparticles, gold nanoparticles and gold nanourchins in darkfield microscopy (top) and in a immuno-dot blot assay (bottom).
Store silver nanoparticles at 2-8°C and protected from light. Do NOT freeze. If frozen, the silver nanoparticles will irreversibly aggregate. Aggregation of silver nanoparticles is indicated by change in color of the solution and an increase in the absorption of light in the red part of the visible spectrum, as detailed in Silver Nanoparticle Properties Tech Note.
When stored as specified, the colloidal silver is stable for at least one year.
Large silver nanoparticles, i.e., above 60 nm in size, might sediment over time but will not affect the performance of the silver nanoparticles. Prior to use, re-suspend the sedimented particles by swirling the container until a homogenous solution is obtained.
For optimal performance and stability of the product, silver nanoparticles should not be stored diluted from the original concentration or in a bottle or flask other than that the product is supplied in. Care should be taken to use clean storage containers if other than supplied with the product.
Although it is not always necessary, washing of silver nanoparticles might be required for certain applications. Silver ions released from the silver nanoparticle surface may affect some applications. The easiest way to remove such possible contaminants from the silver nanoparticle solution is by centrifugation. Centrifugal force is dependent on the size of silver nanoparticles, as listed Table I.
Note:
1. Surfactant should be present during centrifugation, to prevent the nanoparticles from aggregation.Table I.Appropriate G force for the centrifugation of silver nanoparticles. The conditions are based on centrifuging 1 mL of silver nanoparticles in a 1.5 ml microcentrifuge tubes using a standard microcentrifuge.
| Size (nm) | Speed (g) | Time (min) |
|---|---|---|
| 10* | 21,000 | 60 |
| 20 | 17,000 | 30 |
| 30 | 11,000 | 30 |
| 40 | 3,000 | 30 |
| 50 | 1,800 | 30 |
| 60 | 900 | 30 |
| 80 | 500 | 30 |
| 100 | 300 | 30 |
*For 10nm silver nanoparticles, recovery was about 50% at the specified condition. For better recovery, 1) use an ultracentrifuge to achieve higher G force; 2) use a Centricon Spin Column.
Passive adsorption of a protein molecule onto silver nanoparticles is mediated by the electrostatic and hydrophobic interactions between the protein molecule and the surface layer of the colloidal silver. This process is maximally achieved at a pH close to the pI of the protein to be conjugated. Another important parameter is the amount of protein loading. If surface-adsorbed protein is not sufficient, aggregation occurs upon addition of electrolytes present in standard buffers. Therefore, a titration is required to determine the protein concentration for complete saturation and shielding of the silver nanoparticle surface.
However, some proteins may undergo disturbances to the tertiary structure upon passive adsorption, which may impair their affinity and/or specificity in molecular binding applications. To better preserve the activity of the conjugated protein an alternative approach can be used, i.e., covalently coupling protein molecules onto silver nanoparticles functionalized with carboxyl groups via a PEG-linker. The PEG-linker allows for more flexibility and also better accessibility of the conjugated protein to its antigen/substrate due to the inherent mobility of the PEG-linker and the increased distance from the gold surface. Further, the shielding polyethylene glycol (PEG) layer generally also result in conjugates with superior stability and less non-specific binding. Conjugation to these types of functionalized particles can be performed using straightforward carbodiimide (EDC/NHS) coupling chemistry.
Note: The amount of protein needed to saturate the silver colloid can also be determined and verified through agarose gel-electroporesis. Binding of protein to the silver nanoparticle surface changes the overall particle charge and size both of which will affect the migration pattern in the agarose gel.
----------------------
Figure 1.Validation of functionality (biotin binding) of a streptavidin silver nanoparticle conjugate using an immuno-dot blot assay.
-------------
Table I. Recommended centrifugation speeds for silver nanoparticles of different sizes. Note that centrifugation times and speeds are based on a 1 ml sample volume in 1.5 ml microcentrifuge tubes.
| Silver Nanoparticle Diameter (nm) | Centrifugation Force | Time (min) |
| 10nm | 21,000 x g | 60 |
| 20nm | 17,000 x g | 30 |
| 30nm | 11,000 x g | 30 |
| 40nm | 3,000 x g | 30 |
| 50nm | 1,500 x g | 30 |
| 60nm | 900 x g | 30 |
| 70nm | 700 x g | 30 |
| 80nm | 500 x g | 30 |
| 90nm | 400 x g | 30 |
| 100nm | 300 x g | 30 |
Figure 1.
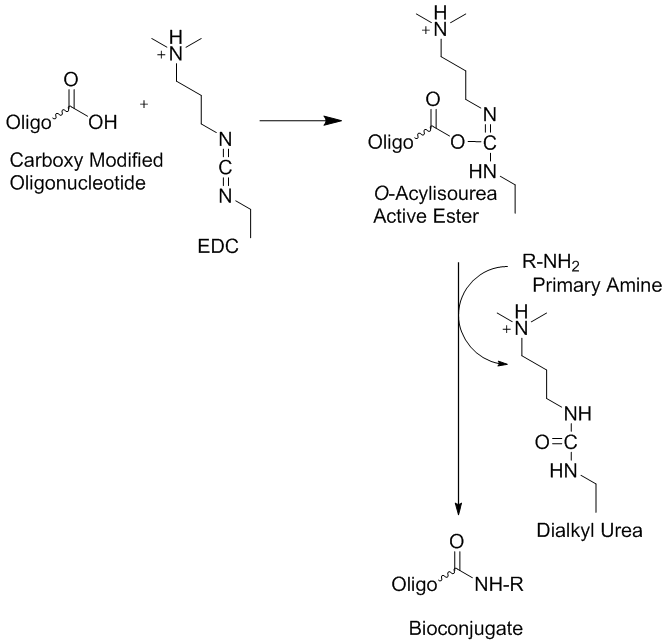

Using that strategy carboxyl modified oligonucleotides can be easily immobilized on solid support such as micro-array slides and various types of aminoalkylated beads.
Bio-synthesis offers not only wide varieties of 3’-, 5’- and internally carboxyl modified oligonucleotides and also their conjugates with peptides, proteins and antibodies.1
References:
1. http://www.biosyn.com/Bioconjugation.aspx
2. E. Jablonski, E. W.Moomaw, R. H.Tullis and J. L.Ruth Nucleic Acid Res., 1986, 14, 6115-6128.
3. J. D. Kahl and M. M. Greenberg J. Org. Chem., 1999, 64 (2), 507–510
4. A.V. Kachalova, T. S. Zatsepin, E. A. Romanova, D. A. Strelenko, M. J. Gait, T. S. Oretskaya Nucleosides, Nucleotides Nucleic Acids. 2000, 19, 1693-1707
5. T. P. Prakash, A. M. Kawasaki, E. A. Lesnik, S. R. Owens, M. Manoharan Org. Lett. 2003, 5, 403-406.